The Babesia bovis VESA1 virulence factor subunit 1b is encoded by the 1beta branch of the ves multigene family
- PMID: 20226217
- PMCID: PMC2856709
- DOI: 10.1016/j.molbiopara.2010.03.001
The Babesia bovis VESA1 virulence factor subunit 1b is encoded by the 1beta branch of the ves multigene family
Abstract
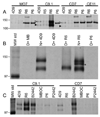
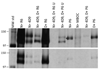
Babesia bovis, an intraerythrocytic parasite of cattle, establishes persistent infections of extreme duration. This is accomplished, at least in part, through rapid antigenic variation of a heterodimeric virulence factor, the variant erythrocyte surface antigen-1 (VESA1) protein. Previously, the VESA1a subunit was demonstrated to be encoded by a 1alpha member of the ves multigene family. Since its discovery the 1beta branch of this multigene family has been hypothesized to encode the VESA1b polypeptide, but formal evidence for this connection has been lacking. Here, we provide evidence that products of ves1beta genes are rapidly variant in antigenicity and size-polymorphic, matching known VESA1b polypeptides. Importantly, the ves1beta-encoded antigens are co-precipitated with VESA1a during immunoprecipitation with anti-VESA1a monoclonal antibodies, and antisera to ves1beta polypeptide co-precipitate VESA1a. Further, the ves1beta-encoded antigens significantly co-localize with VESA1a on the infected-erythrocyte membrane surface of live cells. These characteristics all match known properties of VESA1b, allowing us to conclude that the ves1beta gene divergently apposing the ves1beta gene within the locus of active ves transcription (LAT) encodes the 1b subunit of the VESA1 cytoadhesion ligand. However, the extent and stoichiometry of VESA1a and 1b co-localization on the surface of individual cells is quite variable, implicating competing effects on transcription, translation, or trafficking of the two subunits. These results provide essential information facilitating further investigation into this parasite virulence factor.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Unusual chromatin structure associated with monoparalogous transcription of the Babesia bovis ves multigene family.Int J Parasitol. 2013 Feb;43(2):163-72. doi: 10.1016/j.ijpara.2012.10.018. Epub 2012 Nov 23. Int J Parasitol. 2013. PMID: 23178996 Free PMC article.
-
Characterization of the unusual bidirectional ves promoters driving VESA1 expression and associated with antigenic variation in Babesia bovis.Eukaryot Cell. 2012 Mar;11(3):260-9. doi: 10.1128/EC.05318-11. Epub 2012 Jan 27. Eukaryot Cell. 2012. PMID: 22286091 Free PMC article.
-
Antigenic variation in Babesia bovis occurs through segmental gene conversion of the ves multigene family, within a bidirectional locus of active transcription.Mol Microbiol. 2006 Jan;59(2):402-14. doi: 10.1111/j.1365-2958.2005.04993.x. Mol Microbiol. 2006. PMID: 16390438
-
Antigenic variation as an exploitable weakness of babesial parasites.Vet Parasitol. 2006 May 31;138(1-2):50-60. doi: 10.1016/j.vetpar.2006.01.039. Epub 2006 Mar 3. Vet Parasitol. 2006. PMID: 16517078 Review.
-
Search for Babesia bovis vaccine candidates.Parassitologia. 2007 May;49 Suppl 1:9-12. Parassitologia. 2007. PMID: 17691600 Review.
Cited by
-
Identification of a novel variant erythrocyte surface antigen-1 (VESA1) in Babesia orientalis.Parasitol Res. 2021 Aug;120(8):2863-2872. doi: 10.1007/s00436-021-07194-9. Epub 2021 Jul 5. Parasitol Res. 2021. PMID: 34219188 Free PMC article.
-
Babesia bovis Rad51 ortholog influences switching of ves genes but is not essential for segmental gene conversion in antigenic variation.PLoS Pathog. 2020 Aug 31;16(8):e1008772. doi: 10.1371/journal.ppat.1008772. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32866214 Free PMC article.
-
Variable and Variant Protein Multigene Families in Babesia bovis Persistence.Pathogens. 2019 Jun 11;8(2):76. doi: 10.3390/pathogens8020076. Pathogens. 2019. PMID: 31212587 Free PMC article. Review.
-
ves1α genes expression is the major determinant of Babesia bovis-infected erythrocytes cytoadhesion to endothelial cells.PLoS Pathog. 2025 Apr 28;21(4):e1012583. doi: 10.1371/journal.ppat.1012583. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40294074 Free PMC article.
-
Unusual chromatin structure associated with monoparalogous transcription of the Babesia bovis ves multigene family.Int J Parasitol. 2013 Feb;43(2):163-72. doi: 10.1016/j.ijpara.2012.10.018. Epub 2012 Nov 23. Int J Parasitol. 2013. PMID: 23178996 Free PMC article.
References
-
- Kuttler KL. World-wide impact of babesiosis. In: Ristic M, editor. Babesiosis of Domestic Animals and Man. Boca Raton: CRC Press, Inc.; 1988. pp. 1–22.
-
- Wright IG, Goodger BV, Clark IA. Immunopathophysiology of Babesia bovis and Plasmodium falciparum infections. Parasitol Today. 1988;4:214–218. - PubMed
-
- Wright IG, Goodger BV, Buffington GD, Clark IA, Parrodi F, Waltisbuhl DJ. Immunopathophysiology of babesial infections. Trans R Soc Trop Med Hyg. 1989;83 Supplement:11–13. - PubMed
-
- Jeffery GM, Eyles DE. The duration in the human host of infections with a Panama strain of Plasmodium falciparum. Am J Trop Med Hyg. 1954;2:219–224. - PubMed
-
- Bottius E, Guanzirolli A, Trape JF, Rogier C, Konate L, Druilhe P. Malaria: even more chronic in nature than previously thought; evidence for subpatent parasitemia detectable by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1996;90:15–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

