Single round of antigen receptor signaling programs naive B cells to receive T cell help
- PMID: 20226693
- PMCID: PMC3607434
- DOI: 10.1016/j.immuni.2010.02.013
Single round of antigen receptor signaling programs naive B cells to receive T cell help
Abstract
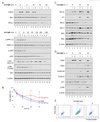
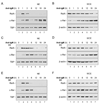
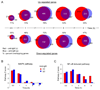
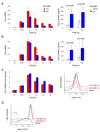
To simulate transient B cell activation that is the likely initiator of T-dependent responses, we examined the molecular and functional consequences of a single round of immunoglobulin M (IgM) signaling. This form of activation triggered early cytosolic signaling and the transcription factor NF-kappaB activation indistinguishably from conventional continuous IgM crosslinking but did not induce G1 progression. However, single round IgM signaling changed the expression of chemokine and chemokine receptor genes implicated in initiating T-dependent responses, as well as accentuated responsiveness to CD40 signaling. Several features of single-round IgM signaling in vitro were recapitulated in B cells after short-term exposure to antigen in vivo. We propose that transient BCR signals prime B cells to receive T cell help by increasing the probability of B-T encounter and creating a cellular environment that is hyper-responsive to CD40 signaling.
Figures

Similar articles
-
Synergistic B cell activation by CD40 and the B cell antigen receptor: role of B lymphocyte antigen receptor-mediated kinase activation and tumor necrosis factor receptor-associated factor regulation.J Biol Chem. 2004 Jan 23;279(4):2575-82. doi: 10.1074/jbc.M310628200. Epub 2003 Nov 6. J Biol Chem. 2004. PMID: 14604983
-
Constitutive expression of the B7h ligand for inducible costimulator on naive B cells is extinguished after activation by distinct B cell receptor and interleukin 4 receptor-mediated pathways and can be rescued by CD40 signaling.J Exp Med. 2002 Jul 1;196(1):97-108. doi: 10.1084/jem.20020298. J Exp Med. 2002. PMID: 12093874 Free PMC article.
-
Cutting edge: CD40 engagement eliminates the need for Bruton's tyrosine kinase in B cell receptor signaling for NF-kappa B.J Immunol. 2003 Mar 15;170(6):2806-10. doi: 10.4049/jimmunol.170.6.2806. J Immunol. 2003. PMID: 12626529
-
Exosome release by primary B cells.Crit Rev Immunol. 2009;29(3):203-17. doi: 10.1615/critrevimmunol.v29.i3.20. Crit Rev Immunol. 2009. PMID: 19538135 Review.
-
Key molecules involved in receptor-mediated lymphocyte activation.Curr Protoc Immunol. 2001 Nov;Chapter 11:Unit 11.9A. doi: 10.1002/0471142735.im1109as44. Curr Protoc Immunol. 2001. PMID: 18432708 Review.
Cited by
-
NR4A nuclear receptors restrain B cell responses to antigen when second signals are absent or limiting.Nat Immunol. 2020 Oct;21(10):1267-1279. doi: 10.1038/s41590-020-0765-7. Epub 2020 Aug 31. Nat Immunol. 2020. PMID: 32868928 Free PMC article.
-
B cell TLRs and induction of immunoglobulin class-switch DNA recombination.Front Biosci (Landmark Ed). 2012 Jun 1;17(7):2594-615. doi: 10.2741/4073. Front Biosci (Landmark Ed). 2012. PMID: 22652800 Free PMC article. Review.
-
Differential requirements for the canonical NF-κB transcription factors c-REL and RELA during the generation and activation of mature B cells.Immunol Cell Biol. 2017 Mar;95(3):261-271. doi: 10.1038/icb.2016.95. Epub 2016 Sep 21. Immunol Cell Biol. 2017. PMID: 27649781 Free PMC article.
-
The NFκB signaling system in the generation of B-cell subsets: from germinal center B cells to memory B cells and plasma cells.Front Immunol. 2023 Dec 11;14:1185597. doi: 10.3389/fimmu.2023.1185597. eCollection 2023. Front Immunol. 2023. PMID: 38169968 Free PMC article. Review.
-
Molecular programming of B cell memory.Nat Rev Immunol. 2011 Dec 9;12(1):24-34. doi: 10.1038/nri3128. Nat Rev Immunol. 2011. PMID: 22158414 Free PMC article. Review.
References
-
- Aiba Y, Yamazaki T, Okada T, Gotoh K, Sanjo H, Ogata M, Kurosaki T. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006;24:259–268. - PubMed
-
- Bystry RS, Aluvihare V, Welch KA, Kallikourdis M, Betz AG. B cells and professional APCs recruit regulatory T cells via CCL4. Nat Immunol. 2001;2:1126–1132. - PubMed
-
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

