Topological frustration in beta alpha-repeat proteins: sequence diversity modulates the conserved folding mechanisms of alpha/beta/alpha sandwich proteins
- PMID: 20226790
- PMCID: PMC2862464
- DOI: 10.1016/j.jmb.2010.03.001
Topological frustration in beta alpha-repeat proteins: sequence diversity modulates the conserved folding mechanisms of alpha/beta/alpha sandwich proteins
Abstract
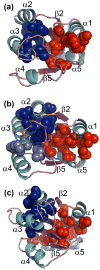
The thermodynamic hypothesis of Anfinsen postulates that structures and stabilities of globular proteins are determined by their amino acid sequences. Chain topology, however, is known to influence the folding reaction, in that motifs with a preponderance of local interactions typically fold more rapidly than those with a larger fraction of nonlocal interactions. Together, the topology and sequence can modulate the energy landscape and influence the rate at which the protein folds to the native conformation. To explore the relationship of sequence and topology in the folding of beta alpha-repeat proteins, which are dominated by local interactions, we performed a combined experimental and simulation analysis on two members of the flavodoxin-like, alpha/beta/alpha sandwich fold. Spo0F and the N-terminal receiver domain of NtrC (NT-NtrC) have similar topologies but low sequence identity, enabling a test of the effects of sequence on folding. Experimental results demonstrated that both response-regulator proteins fold via parallel channels through highly structured submillisecond intermediates before accessing their cis prolyl peptide bond-containing native conformations. Global analysis of the experimental results preferentially places these intermediates off the productive folding pathway. Sequence-sensitive Gō-model simulations conclude that frustration in the folding in Spo0F, corresponding to the appearance of the off-pathway intermediate, reflects competition for intra-subdomain van der Waals contacts between its N- and C-terminal subdomains. The extent of transient, premature structure appears to correlate with the number of isoleucine, leucine, and valine (ILV) side chains that form a large sequence-local cluster involving the central beta-sheet and helices alpha2, alpha 3, and alpha 4. The failure to detect the off-pathway species in the simulations of NT-NtrC may reflect the reduced number of ILV side chains in its corresponding hydrophobic cluster. The location of the hydrophobic clusters in the structure may also be related to the differing functional properties of these response regulators. Comparison with the results of previous experimental and simulation analyses on the homologous CheY argues that prematurely folded unproductive intermediates are a common property of the beta alpha-repeat motif.
(c) 2010 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Modulation of frustration in folding by sequence permutation.Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):10562-7. doi: 10.1073/pnas.1324230111. Epub 2014 Jul 7. Proc Natl Acad Sci U S A. 2014. PMID: 25002512 Free PMC article.
-
Kinetic traps in the folding of beta alpha-repeat proteins: CheY initially misfolds before accessing the native conformation.J Mol Biol. 2008 Oct 3;382(2):467-84. doi: 10.1016/j.jmb.2008.06.054. Epub 2008 Jun 28. J Mol Biol. 2008. PMID: 18619461
-
Subdomain competition, cooperativity, and topological frustration in the folding of CheY.J Mol Biol. 2008 Oct 3;382(2):485-95. doi: 10.1016/j.jmb.2008.07.007. Epub 2008 Jul 11. J Mol Biol. 2008. PMID: 18644380 Free PMC article.
-
Folding of proteins with a flavodoxin-like architecture.FEBS J. 2017 Oct;284(19):3145-3167. doi: 10.1111/febs.14077. Epub 2017 Apr 28. FEBS J. 2017. PMID: 28380286 Review.
-
Structural conservation in the CheY superfamily.Biochemistry. 1993 Nov 9;32(44):11741-53. doi: 10.1021/bi00095a001. Biochemistry. 1993. PMID: 8218244 Review. No abstract available.
Cited by
-
Modulation of frustration in folding by sequence permutation.Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):10562-7. doi: 10.1073/pnas.1324230111. Epub 2014 Jul 7. Proc Natl Acad Sci U S A. 2014. PMID: 25002512 Free PMC article.
-
Take home lessons from studies of related proteins.Curr Opin Struct Biol. 2013 Feb;23(1):66-74. doi: 10.1016/j.sbi.2012.11.009. Epub 2012 Dec 20. Curr Opin Struct Biol. 2013. PMID: 23265640 Free PMC article. Review.
-
Understanding the folding-function tradeoff in proteins.PLoS One. 2013 Apr 12;8(4):e61222. doi: 10.1371/journal.pone.0061222. Print 2013. PLoS One. 2013. PMID: 23593437 Free PMC article.
-
Energetic frustrations in protein folding at residue resolution: a homologous simulation study of Im9 proteins.PLoS One. 2014 Jan 31;9(1):e87719. doi: 10.1371/journal.pone.0087719. eCollection 2014. PLoS One. 2014. PMID: 24498176 Free PMC article.
-
Sequence-, structure-, and dynamics-based comparisons of structurally homologous CheY-like proteins.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1578-1583. doi: 10.1073/pnas.1621344114. Epub 2017 Jan 31. Proc Natl Acad Sci U S A. 2017. PMID: 28143938 Free PMC article.
References
-
- Anfinsen CB, Scheraga HA. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. - PubMed
-
- Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. - PubMed
-
- Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. - PubMed
-
- Kamagata K, Kuwajima K. Surprisingly high correlation between early and late stages in non-two-state protein folding. J Mol Biol. 2006;357:1647–1654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

