Circadian clock control of the cellular response to DNA damage
- PMID: 20227409
- PMCID: PMC2878924
- DOI: 10.1016/j.febslet.2010.03.017
Circadian clock control of the cellular response to DNA damage
Abstract
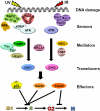
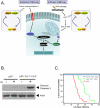
Mammalian cells possess a cell-autonomous molecular clock which controls the timing of many biochemical reactions and hence the cellular response to environmental stimuli including genotoxic stress. The clock consists of an autoregulatory transcription-translation feedback loop made up of four genes/proteins, BMal1, Clock, Cryptochrome, and Period. The circadian clock has an intrinsic period of about 24 h, and it dictates the rates of many biochemical reactions as a function of the time of the day. Recently, it has become apparent that the circadian clock plays an important role in determining the strengths of cellular responses to DNA damage including repair, checkpoints, and apoptosis. These new insights are expected to guide development of novel mechanism-based chemotherapeutic regimens.
Copyright 2010 Federation of European Biochemical Societies. All rights reserved.
Figures




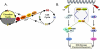
References
-
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. - PubMed
-
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. - PubMed
-
- Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

