The Longus type IV pilus of enterotoxigenic Escherichia coli (ETEC) mediates bacterial self-aggregation and protection from antimicrobial agents
- PMID: 20227481
- PMCID: PMC2860038
- DOI: 10.1016/j.micpath.2010.03.006
The Longus type IV pilus of enterotoxigenic Escherichia coli (ETEC) mediates bacterial self-aggregation and protection from antimicrobial agents
Abstract
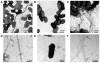
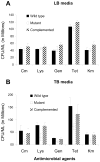
Enterotoxigenic Escherichia coli (ETEC) strains are leading causes of childhood diarrhea in developing countries. ETEC pili and non-pili adherence factors designated colonization surface antigens (CSA) are believed to be important in the pathogenesis of diarrhea. Longus, a type IV pilus identified as the CSA(21), is expressed in up to one-third of ETEC strains, and share similarities to the toxin-coregulated pilus of Vibrio cholerae, and the bundle-forming pilus of enteropathogenic E. coli. To identify longus phenotype and possible function, a site-directed mutation of the lngA major subunit gene in the E9034A wild type ETEC strain was constructed. Lack of longus expression from the lngA mutant was demonstrated by immunoblot analysis and electron microscopy using specific anti-LngA antibody. Formation of self-aggregates by ETEC was shown to be dependent on longus expression as the lngA mutant or wild type grown under poor longus expression conditions was unable to express this phenotype. Longus-expressing ETEC were also associated with improved survival when exposed to antibacterial factors including lysozyme and antibiotics. This suggests that longus-mediated bacterial self-aggregates protect bacteria against antimicrobial environmental agents and may promote gut colonization.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Identification of lngA, the structural gene of longus type IV pilus of enterotoxigenic Escherichia coli.Microbiology (Reading). 1999 Jul;145 ( Pt 7):1809-1816. doi: 10.1099/13500872-145-7-1809. Microbiology (Reading). 1999. PMID: 10439420
-
Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli.Mol Microbiol. 1994 Apr;12(1):71-82. doi: 10.1111/j.1365-2958.1994.tb00996.x. Mol Microbiol. 1994. PMID: 7914665
-
Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells.J Bacteriol. 2010 Jun;192(11):2791-800. doi: 10.1128/JB.01595-09. Epub 2010 Mar 26. J Bacteriol. 2010. PMID: 20348256 Free PMC article.
-
Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili--a minireview.Gene. 1997 Jun 11;192(1):39-43. doi: 10.1016/s0378-1119(97)00039-5. Gene. 1997. PMID: 9224872 Review.
-
Strategies to overexpress enterotoxigenic Escherichia coli (ETEC) colonization factors for the construction of oral whole-cell inactivated ETEC vaccine candidates.Appl Microbiol Biotechnol. 2012 Mar;93(6):2291-300. doi: 10.1007/s00253-012-3930-6. Epub 2012 Feb 16. Appl Microbiol Biotechnol. 2012. PMID: 22350259 Review.
Cited by
-
Recombinant Escherichia coli BL21 with LngA Variants from ETEC E9034A Promotes Adherence to HT-29 Cells.Pathogens. 2023 Feb 16;12(2):337. doi: 10.3390/pathogens12020337. Pathogens. 2023. PMID: 36839609 Free PMC article.
-
Recent advances in understanding enteric pathogenic Escherichia coli.Clin Microbiol Rev. 2013 Oct;26(4):822-80. doi: 10.1128/CMR.00022-13. Clin Microbiol Rev. 2013. PMID: 24092857 Free PMC article. Review.
-
CS21 positive multidrug-resistant ETEC clinical isolates from children with diarrhea are associated with self-aggregation, and adherence.Front Microbiol. 2014 Dec 17;5:709. doi: 10.3389/fmicb.2014.00709. eCollection 2014. Front Microbiol. 2014. PMID: 25646093 Free PMC article.
-
Enterotoxigenic Escherichia coli CS21 pilus contributes to adhesion to intestinal cells and to pathogenesis under in vivo conditions.Microbiology (Reading). 2013 Aug;159(Pt 8):1725-1735. doi: 10.1099/mic.0.065532-0. Epub 2013 Jun 12. Microbiology (Reading). 2013. PMID: 23760820 Free PMC article.
-
Horizontal gene transfers with or without cell fusions in all categories of the living matter.Adv Exp Med Biol. 2011;714:5-89. doi: 10.1007/978-94-007-0782-5_2. Adv Exp Med Biol. 2011. PMID: 21506007 Free PMC article. Review.
References
-
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
-
- Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–8. - PubMed
-
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. - PubMed
-
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–22. - PubMed
-
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

