Agrobacterium induces expression of a host F-box protein required for tumorigenicity
- PMID: 20227663
- PMCID: PMC3427693
- DOI: 10.1016/j.chom.2010.02.009
Agrobacterium induces expression of a host F-box protein required for tumorigenicity
Abstract
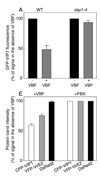
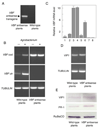
Agrobacterium exports DNA into plant cells, eliciting neoplastic growths on many plant species. During this process, a Skp1-Cdc53-cullin-F-box (SCF) complex that contains the bacterial virulence F-box protein VirF facilitates genetic transformation by targeting for proteolysis proteins, the Agrobacterium protein VirE2 and the host protein VIP1, that coat the transferred DNA. However, some plant species do not require VirF for transformation. Here, we show that Agrobacterium induces expression of a plant F-box protein, which we designated VBF for VIP1-binding F-box protein, that can functionally replace VirF, regulating levels of the VirE2 and VIP1 proteins via a VBF-containing SCF complex. When expressed in Agrobacterium and exported into the plant cell, VBF functionally complements tumor formation by a strain lacking VirF. VBF expression is known to be induced by diverse pathogens, suggesting that Agrobacterium has co-opted a plant defense response and that bacterial VirF and plant VBF both contribute to targeted proteolysis that promotes plant genetic transformation.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium.Nature. 2004 Sep 2;431(7004):87-92. doi: 10.1038/nature02857. Nature. 2004. PMID: 15343337
-
Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein.Curr Biol. 2001 Feb 20;11(4):258-62. doi: 10.1016/s0960-9822(01)00069-0. Curr Biol. 2001. PMID: 11250154
-
The putative Agrobacterium transcriptional activator-like virulence protein VirD5 may target T-complex to prevent the degradation of coat proteins in the plant cell nucleus.New Phytol. 2014 Sep;203(4):1266-1281. doi: 10.1111/nph.12866. Epub 2014 May 28. New Phytol. 2014. PMID: 24865527
-
Finding a way to the nucleus.Curr Opin Microbiol. 2010 Feb;13(1):53-8. doi: 10.1016/j.mib.2009.11.003. Epub 2009 Dec 21. Curr Opin Microbiol. 2010. PMID: 20022799 Review.
-
The role of the ubiquitin-proteasome system in Agrobacterium tumefaciens-mediated genetic transformation of plants.Plant Physiol. 2012 Sep;160(1):65-71. doi: 10.1104/pp.112.200949. Epub 2012 Jul 10. Plant Physiol. 2012. PMID: 22786890 Free PMC article. Review. No abstract available.
Cited by
-
A plasmodesmal glycosyltransferase-like protein.PLoS One. 2013;8(2):e58025. doi: 10.1371/journal.pone.0058025. Epub 2013 Feb 26. PLoS One. 2013. PMID: 23469135 Free PMC article.
-
The tomato yellow leaf curl virus (TYLCV) V2 protein interacts with the host papain-like cysteine protease CYP1.Plant Signal Behav. 2012 Aug;7(8):983-9. doi: 10.4161/psb.20935. Epub 2012 Jul 25. Plant Signal Behav. 2012. PMID: 22827939 Free PMC article.
-
Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins.Cell Microbiol. 2015 Jan;17(1):18-25. doi: 10.1111/cmi.12385. Epub 2014 Nov 25. Cell Microbiol. 2015. PMID: 25339602 Free PMC article. Review.
-
Assaying proteasomal degradation in a cell-free system in plants.J Vis Exp. 2014 Mar 26;(85):51293. doi: 10.3791/51293. J Vis Exp. 2014. PMID: 24747194 Free PMC article.
-
TRA1: A Locus Responsible for Controlling Agrobacterium-Mediated Transformability in Barley.Front Plant Sci. 2020 Apr 16;11:355. doi: 10.3389/fpls.2020.00355. eCollection 2020. Front Plant Sci. 2020. PMID: 32373138 Free PMC article.
References
-
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. - PubMed
-
- Bartel P, Chien C, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

