A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues
- PMID: 20228232
- PMCID: PMC2879958
- DOI: 10.1124/dmd.109.031831
A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues
Abstract
HepaRG cells, derived from a female hepatocarcinoma patient, are capable of differentiating into biliary epithelial cells and hepatocytes. More importantly, differentiated HepaRG cells are able to maintain activities of many xenobiotic-metabolizing enzymes, and expression of the metabolizing enzyme genes can be induced by xenobiotics. The ability of these cells to express and induce xenobiotic-metabolizing enzymes is in stark contrast to the frequently used HepG2 cells. The previous studies have mainly focused on a set of selected genes; therefore, it is of significant interest to know the extent of similarity of gene expression at whole genome levels in HepaRG cells and HepG2 cells compared with primary human hepatocytes and human liver tissues. To accomplish this objective, we used Affymetrix (Santa Clara, CA) U133 Plus 2.0 arrays to characterize the whole genome gene expression profiles in triplicate biological samples from HepG2 cells, HepaRG cells (undifferentiated and differentiated cells), freshly isolated primary human hepatocytes, and frozen liver tissues. After using similarity matrix, principal components, and hierarchical clustering methods, we found that HepaRG cells globally transcribe genes at levels more similar to human primary hepatocytes and human liver tissues than HepG2 cells. In particular, many genes encoding drug-processing proteins are transcribed at a more similar level in HepaRG cells than in HepG2 cells compared with primary human hepatocytes and liver samples. The transcriptomic similarity of HepaRG with primary human hepatocytes is encouraging for use of HepaRG cells in the study of xenobiotic metabolism, hepatotoxicology, and hepatocyte differentiation.
Figures
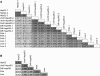


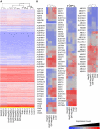

References
-
- Aninat C, Piton A, Glaise D, Le Charpentier T, Langouët S, Morel F, Guguen-Guillouzo C, Guillouzo A. (2006) Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34:75–83 - PubMed
-
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3 - PubMed
-
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. (2007) The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168:66–73 - PubMed
-
- Hewitt NJ, Lechón MJ, Houston JB, Hallifax D, Brown HS, Maurel P, Kenna JG, Gustavsson L, Lohmann C, Skonberg C, et al. (2007) Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev 39:159–234 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

