High expression of lymphocyte-activation gene 3 (LAG3) in chronic lymphocytic leukemia cells is associated with unmutated immunoglobulin variable heavy chain region (IGHV) gene and reduced treatment-free survival
- PMID: 20228263
- PMCID: PMC2860469
- DOI: 10.2353/jmoldx.2010.090100
High expression of lymphocyte-activation gene 3 (LAG3) in chronic lymphocytic leukemia cells is associated with unmutated immunoglobulin variable heavy chain region (IGHV) gene and reduced treatment-free survival
Abstract
Chronic lymphocytic leukemia (CLL) is characterized by a monoclonal expansion of mature B-lymphocytes. Mutational status of the immunoglobulin variable heavy chain region (IGHV) gene stratifies CLL patients into two prognostic groups. We performed microarray analysis of CLL cells using the Agilent platform to detect the most important gene expression differences regarding IGHV status in CLL cells. We analyzed a cohort of 118 CLL patients with different IGHV mutational status and completely characterized all described prognostic markers using expression microarrays and quantitative real-time RT-PCR (reverse transcription PCR). We detected lymphocyte-activation gene 3 (LAG3) as a novel prognostic marker: LAG3 high expression in CLL cells correlates with unmutated IGHV (P < 0.0001) and reduced treatment-free survival (P = 0.0087). Furthermore, quantitative real-time RT-PCR analysis identified a gene-set (LAG3, LPL, ZAP70) whose overexpression is assigned to unmutated IGHV with 90% specificity (P < 0.0001). Moreover, high expression of tested gene-set and unmutated IGHV equally correlated with reduced treatment-free survival (P = 7.7 * 10(-11) vs. P = 1.8 * 10(-11)). Our results suggest that IGHV status can be precisely assessed using the expression analysis of LAG3, LPL, and ZAP70 genes. Expression data of tested markers provides a similar statistical concordance with treatment-free survival as that of the IGHV status itself. Our findings contribute to the elucidation of CLL pathogenesis and provide novel prognostic markers for possible application in routine diagnostics.
Figures
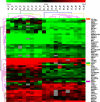
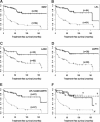
Similar articles
-
COBLL1, LPL and ZAP70 expression defines prognostic subgroups of chronic lymphocytic leukemia patients with high accuracy and correlates with IGHV mutational status.Leuk Lymphoma. 2017 Jan;58(1):70-79. doi: 10.1080/10428194.2016.1180690. Epub 2016 May 17. Leuk Lymphoma. 2017. PMID: 27185377
-
Mutational status and gene repertoire of IGHV-IGHD-IGHJ rearrangements in Serbian patients with chronic lymphocytic leukemia.Clin Lymphoma Myeloma Leuk. 2012 Aug;12(4):252-60. doi: 10.1016/j.clml.2012.03.005. Epub 2012 May 4. Clin Lymphoma Myeloma Leuk. 2012. PMID: 22560084
-
ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile.Blood. 2003 Jun 15;101(12):4944-51. doi: 10.1182/blood-2002-10-3306. Epub 2003 Feb 20. Blood. 2003. PMID: 12595313
-
Why Is the Immunoglobulin Heavy Chain Gene Mutation Status a Prognostic Indicator in Chronic Lymphocytic Leukemia?Acta Haematol. 2018;140(1):51-54. doi: 10.1159/000491382. Epub 2018 Aug 16. Acta Haematol. 2018. PMID: 30114695 Free PMC article. Review.
-
[Biological characteristics and clinical significance of stereotyped B-cell receptor in chronic lymphocytic leukemia].Zhonghua Xue Ye Xue Za Zhi. 2024 Feb 14;45(2):197-202. doi: 10.3760/cma.j.cn121090-20230718-00012. Zhonghua Xue Ye Xue Za Zhi. 2024. PMID: 38604800 Free PMC article. Review. Chinese.
Cited by
-
LAG3 (CD223) as a cancer immunotherapy target.Immunol Rev. 2017 Mar;276(1):80-96. doi: 10.1111/imr.12519. Immunol Rev. 2017. PMID: 28258692 Free PMC article. Review.
-
Super-Enhancer-Associated nine-gene prognostic score model for prediction of survival in chronic lymphocytic leukemia patients.Front Genet. 2022 Sep 15;13:1001364. doi: 10.3389/fgene.2022.1001364. eCollection 2022. Front Genet. 2022. PMID: 36186463 Free PMC article.
-
LAG-3 expression on tumor-infiltrating T cells in soft tissue sarcoma correlates with poor survival.Cancer Biol Med. 2019 May;16(2):331-340. doi: 10.20892/j.issn.2095-3941.2018.0306. Cancer Biol Med. 2019. PMID: 31516753 Free PMC article.
-
Molecular Pathways and Mechanisms of LAG3 in Cancer Therapy.Clin Cancer Res. 2022 Dec 1;28(23):5030-5039. doi: 10.1158/1078-0432.CCR-21-2390. Clin Cancer Res. 2022. PMID: 35579997 Free PMC article.
-
Immunomodulation in leukemia: cellular aspects of anti-leukemic properties.Clin Transl Oncol. 2020 Jan;22(1):1-10. doi: 10.1007/s12094-019-02132-9. Epub 2019 May 24. Clin Transl Oncol. 2020. PMID: 31127471 Review.
References
-
- Küppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–1529. - PubMed
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. - PubMed
-
- Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234. - PubMed
-
- Binet JL, Lepoprier M, Dighiero G, Charron D, D'Athis P, Vaugier G, Beral HM, Natali JC, Raphael M, Nizet B, Follezou JY. A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer. 1977;40:855–864. - PubMed
-
- Döhner H, Stilgenbauer S, Döhner K, Bentz M, Lichter P. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J Mol Med. 1999;77:266–281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

