Validation and comparison of pharmacogenetics-based warfarin dosing algorithms for application of pharmacogenetic testing
- PMID: 20228265
- PMCID: PMC2860463
- DOI: 10.2353/jmoldx.2010.090110
Validation and comparison of pharmacogenetics-based warfarin dosing algorithms for application of pharmacogenetic testing
Abstract
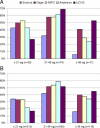
Warfarin is a widely prescribed drug that is difficult to use because of its narrow therapeutic window. Genetic polymorphisms associated with warfarin metabolism have been identified, but the clinical utility of genetic testing in warfarin dosing has not been established. External validation of published algorithms is critical to determine the best prediction for warfarin dosing in prospective trials. We used two independent datasets totaling 1095 patients to evaluate four published algorithms and a simple prediction algorithm developed in this study based on the CYP2C9*2, CYP2C9*3, and VKORC1 -1639 polymorphisms in 150 patients taking warfarin. Predicted warfarin doses were calculated and compared for accuracy with actual maintenance doses. All evaluated pharmacogenetics-based dosing algorithms performed similarly for both datasets. The proportion of variation explained (R(2)) was high (60% to 65%) in the small white-only Connecticut dataset but low (36% to 46%) in the large dataset on a diverse ethnic population from the International Warfarin Pharmacogenetics Consortium (IWPC). When comparing the percentage of patients whose predicted dosage are within 20% of actual, the IWPC algorithm performed the best overall (45.9%) for the two datasets combined while other algorithms performed nearly as well. Because no algorithm could be considered the best for all dosing ranges, it may be important to consider the nature of a local service population in choosing the most appropriate pharmacogenetics-based dosing algorithm.
Figures




References
-
- Jones M, McEwan P, Morgan CL, Peters JR, Goodfellow J, Currie CJ. Evaluation of the pattern of treatment, level of anticoagulation control, and outcome of treatment with warfarin in patients with non-valvar atrial fibrillation: a record linkage study in a large British population. Heart. 2005;91:472–477. - PMC - PubMed
-
- Linkins LA, Choi PT, Douketis JD. Clinical impact of bleeding in patients taking oral anticoagulant therapy for venous thromboembolism: a meta-analysis. Ann Intern Med. 2003;139:893–900. - PubMed
-
- Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7:99–111. - PubMed
-
- Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

