Arabidopsis thaliana as a model organism in systems biology
- PMID: 20228888
- PMCID: PMC2836806
- DOI: 10.1002/wsbm.25
Arabidopsis thaliana as a model organism in systems biology
Abstract
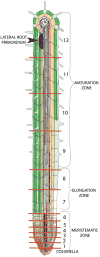
Significant progress has been made in identification of genes and gene networks involved in key biological processes. Yet, how these genes and networks are coordinated over increasing levels of biological complexity, from cells to tissues to organs, remains unclear. To address complex biological questions, biologists are increasingly using high-throughput tools and systems biology approaches to examine complex biological systems at a global scale. A system is a network of interacting and interdependent components that shape the system's unique properties. Systems biology studies the organization of system components and their interactions, with the idea that unique properties of that system can be observed only through study of the system as a whole. The application of systems biology approaches to questions in plant biology has been informative. In this review, we give examples of how systems biology is currently being used in Arabidopsis to investigate the transcriptional networks regulating root development, the metabolic response to stress, and the genetic regulation of metabolic variability. From these studies, we are beginning obtain sufficient data to generate more accurate models for system function. Further investigation of plant systems will require data gathering from specific cells and tissues, continued improvement in metabolic technologies, and novel computational methods for data visualization and modeling.
Keywords: Root development; high-throughput methods; metabolism; quantitative traits; transcriptional networks.
Figures


Similar articles
-
Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks.Plant Cell. 2012 Oct;24(10):3859-75. doi: 10.1105/tpc.112.100776. Epub 2012 Oct 30. Plant Cell. 2012. PMID: 23110892 Free PMC article. Review.
-
Gene regulatory networks in the Arabidopsis root.Curr Opin Plant Biol. 2013 Feb;16(1):50-5. doi: 10.1016/j.pbi.2012.10.007. Epub 2012 Nov 26. Curr Opin Plant Biol. 2013. PMID: 23196272 Review.
-
Genetic control of root growth: from genes to networks.Ann Bot. 2016 Jan;117(1):9-24. doi: 10.1093/aob/mcv160. Epub 2015 Nov 11. Ann Bot. 2016. PMID: 26558398 Free PMC article. Review.
-
Computational systems biology of cellular processes in Arabidopsis thaliana: an overview.Cell Mol Life Sci. 2020 Feb;77(3):433-440. doi: 10.1007/s00018-019-03379-9. Epub 2019 Nov 25. Cell Mol Life Sci. 2020. PMID: 31768604 Free PMC article. Review.
-
Root systems biology: integrative modeling across scales, from gene regulatory networks to the rhizosphere.Plant Physiol. 2013 Dec;163(4):1487-503. doi: 10.1104/pp.113.227215. Epub 2013 Oct 18. Plant Physiol. 2013. PMID: 24143806 Free PMC article. Review.
Cited by
-
Editorial: Model organisms in plant science: Arabidopsis thaliana.Front Plant Sci. 2023 Sep 11;14:1279230. doi: 10.3389/fpls.2023.1279230. eCollection 2023. Front Plant Sci. 2023. PMID: 37767288 Free PMC article. No abstract available.
-
In silico metabolic network analysis of Arabidopsis leaves.BMC Syst Biol. 2016 Oct 29;10(1):102. doi: 10.1186/s12918-016-0347-3. BMC Syst Biol. 2016. PMID: 27793154 Free PMC article.
-
Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues.Plant Cell. 2011 Apr;23(4):1264-71. doi: 10.1105/tpc.110.082867. Epub 2011 Apr 12. Plant Cell. 2011. PMID: 21487096 Free PMC article.
-
NMR spectroscopy analysis reveals differential metabolic responses in arabidopsis roots and leaves treated with a cytokinesis inhibitor.PLoS One. 2020 Nov 6;15(11):e0241627. doi: 10.1371/journal.pone.0241627. eCollection 2020. PLoS One. 2020. PMID: 33156865 Free PMC article.
-
Metabolic Profiling of Intact Arabidopsis thaliana Leaves during Circadian Cycle Using 1H High Resolution Magic Angle Spinning NMR.PLoS One. 2016 Sep 23;11(9):e0163258. doi: 10.1371/journal.pone.0163258. eCollection 2016. PLoS One. 2016. PMID: 27662620 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

