Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii
- PMID: 20230484
- PMCID: PMC2918638
- DOI: 10.1111/j.1467-7652.2010.00503.x
Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii
Abstract
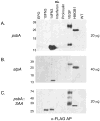
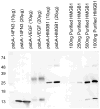
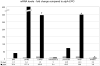
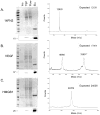
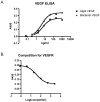
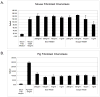
Recombinant proteins are widely used today in many industries, including the biopharmaceutical industry, and can be expressed in bacteria, yeasts, mammalian and insect cell cultures, or in transgenic plants and animals. In addition, transgenic algae have also been shown to support recombinant protein expression, both from the nuclear and chloroplast genomes. However, to date, there are only a few reports on recombinant proteins expressed in the algal chloroplast. It is unclear whether this is because of few attempts or of limitations of the system that preclude expression of many proteins. Thus, we sought to assess the versatility of transgenic algae as a recombinant protein production platform. To do this, we tested whether the algal chloroplast could support the expression of a diverse set of current or potential human therapeutic proteins. Of the seven proteins chosen, >50% expressed at levels sufficient for commercial production. Three expressed at 2%-3% of total soluble protein, while a forth protein accumulated to similar levels when translationally fused to a well-expressed serum amyloid protein. All of the algal chloroplast-expressed proteins are soluble and showed biological activity comparable to that of the same proteins expressed using traditional production platforms. Thus, the success rate, expression levels, and bioactivity achieved demonstrate the utility of Chlamydomonas reinhardtii as a robust platform for human therapeutic protein production.
Figures


Similar articles
-
Accumulation and processing of a recombinant protein designed as a cleavable fusion to the endogenous Rubisco LSU protein in Chlamydomonas chloroplast.BMC Biotechnol. 2009 Mar 26;9:26. doi: 10.1186/1472-6750-9-26. BMC Biotechnol. 2009. PMID: 19323825 Free PMC article.
-
A novel expression platform for the production of diabetes-associated autoantigen human glutamic acid decarboxylase (hGAD65).BMC Biotechnol. 2008 Nov 17;8:87. doi: 10.1186/1472-6750-8-87. BMC Biotechnol. 2008. PMID: 19014643 Free PMC article.
-
Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast.Plant Biotechnol J. 2007 May;5(3):402-12. doi: 10.1111/j.1467-7652.2007.00249.x. Epub 2007 Mar 15. Plant Biotechnol J. 2007. PMID: 17359495
-
Outlook in the application of Chlamydomonas reinhardtii chloroplast as a platform for recombinant protein production.Biotechnol Genet Eng Rev. 2016 Apr-Oct;32(1-2):92-106. doi: 10.1080/02648725.2017.1307673. Epub 2017 Mar 30. Biotechnol Genet Eng Rev. 2016. PMID: 28359189 Review.
-
Recombinant Protein Production in Microalgae: Emerging Trends.Protein Pept Lett. 2020;27(2):105-110. doi: 10.2174/0929866526666191014124855. Protein Pept Lett. 2020. PMID: 31622194 Review.
Cited by
-
Quality Control of Widely Used Therapeutic Recombinant Proteins by a Novel Real-Time PCR Approach.Iran Biomed J. 2016;20(1):56-62. doi: 10.7508/ibj.2016.01.008. Epub 2015 Jun 6. Iran Biomed J. 2016. PMID: 26047906 Free PMC article.
-
Heterologous Gene Expression System Using the Cold-Inducible CnAFP Promoter in Chlamydomonas reinhardtii.J Microbiol Biotechnol. 2020 Nov 28;30(11):1777-1784. doi: 10.4014/jmb.2007.07024. J Microbiol Biotechnol. 2020. PMID: 32807760 Free PMC article.
-
CRISPR-Cas9 System for Genome Engineering of Photosynthetic Microalgae.Mol Biotechnol. 2019 Aug;61(8):541-561. doi: 10.1007/s12033-019-00185-3. Mol Biotechnol. 2019. PMID: 31140149 Review.
-
Towards green biomanufacturing of high-value recombinant proteins using promising cell factory: Chlamydomonas reinhardtii chloroplast.Bioresour Bioprocess. 2022 Aug 13;9(1):83. doi: 10.1186/s40643-022-00568-6. Bioresour Bioprocess. 2022. PMID: 38647750 Free PMC article. Review.
-
Nutritional Health Connection of Algae and its Pharmaceutical Value as Anticancer and Antioxidant Constituents of Drugs.Recent Pat Biotechnol. 2025;19(1):19-34. doi: 10.2174/0118722083287672240321081428. Recent Pat Biotechnol. 2025. PMID: 39840409 Review.
References
-
- Alzari PM, Berglund H, Berrow NS, Blagova E, Busso D, Cambillau C, Campanacci V, Christodoulou E, Eiler S, Fogg MJ, Folkers G, Geerlof A, Hart D, Haouz A, Herman MD, Macieira S, Nordlund P, Perrakis A, Quevillon-Cheruel S, Tarandeau F, van Tilbeurgh H, Unger T, Luna-Vargas MP, Velarde M, Willmanns M, Owens RJ. Implementation of semi-automated cloning and prokaryotic expression screening: the impact of SPINE. Acta Crystallogr D Biol Crystallogr. 2006;62:1103–1113. - PubMed
-
- Arakawa T, Yu J, Chong DK, Hough J, Engen PC, Langridge WH. A plant-based cholera toxin B subunit-insulin fusion protein protects against the development of autoimmune diabetes. Nat Biotechnol. 1998;16:934–938. - PubMed
-
- Aricescu AR, Assenberg R, Bill RM, Busso D, Chang VT, Davis SJ, Dubrovsky A, Gustafsson L, Hedfalk K, Heinemann U, Jones IM, Ksiazek D, Lang C, Maskos K, Messerschmidt A, Macieira S, Peleg Y, Perrakis A, Poterszman A, Schneider G, Sixma TK, Sussman JL, Sutton G, Tarboureich N, Zeev-Ben-Mordehai T, Jones EY. Eukaryotic expression: developments for structural proteomics. Acta Crystallogr D Biol Crystallogr. 2006;62:1114–1124. - PMC - PubMed
-
- Azhagiri AK, Maliga P. Exceptional paternal inheritance of plastids in Arabidopsis suggests that low-frequency leakage of plastids via pollen may be universal in plants. Plant J. 2007;52:817–823. - PubMed
-
- Banci L, Bertini I, Cusack S, de Jong RN, Heinemann U, Jones EY, Kozielski F, Maskos K, Messerschmidt A, Owens R, Perrakis A, Poterszman A, Schneider G, Siebold C, Silman I, Sixma T, Stewart-Jones G, Sussman JL, Thierry JC, Moras D. First steps towards effective methods in exploiting high-throughput technologies for the determination of human protein structures of high biomedical value. Acta Crystallogr D Biol Crystallogr. 2006;62:1208–1217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

