Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes
- PMID: 20230605
- PMCID: PMC2864571
- DOI: 10.1186/gb-2010-11-3-r31
Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes
Abstract
Background: Structured noncoding RNAs perform many functions that are essential for protein synthesis, RNA processing, and gene regulation. Structured RNAs can be detected by comparative genomics, in which homologous sequences are identified and inspected for mutations that conserve RNA secondary structure.
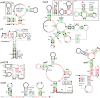
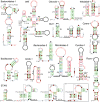
Results: By applying a comparative genomics-based approach to genome and metagenome sequences from bacteria and archaea, we identified 104 candidate structured RNAs and inferred putative functions for many of these. Twelve candidate metabolite-binding RNAs were identified, three of which were validated, including one reported herein that binds the coenzyme S-adenosylmethionine. Newly identified cis-regulatory RNAs are implicated in photosynthesis or nitrogen regulation in cyanobacteria, purine and one-carbon metabolism, stomach infection by Helicobacter, and many other physiological processes. A candidate riboswitch termed crcB is represented in both bacteria and archaea. Another RNA motif may control gene expression from 3'-untranslated regions of mRNAs, which is unusual for bacteria. Many noncoding RNAs that likely act in trans are also revealed, and several of the noncoding RNA candidates are found mostly or exclusively in metagenome DNA sequences.
Conclusions: This work greatly expands the variety of highly structured noncoding RNAs known to exist in bacteria and archaea and provides a starting point for biochemical and genetic studies needed to validate their biologic functions. Given the sustained rate of RNA discovery over several similar projects, we expect that far more structured RNAs remain to be discovered from bacterial and archaeal organisms.
Figures

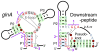

Similar articles
-
The amazing world of bacterial structured RNAs.Genome Biol. 2010;11(3):108. doi: 10.1186/gb-2010-11-3-108. Epub 2010 Mar 15. Genome Biol. 2010. PMID: 20236470 Free PMC article. Review.
-
Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline.Nucleic Acids Res. 2007;35(14):4809-19. doi: 10.1093/nar/gkm487. Epub 2007 Jul 9. Nucleic Acids Res. 2007. PMID: 17621584 Free PMC article.
-
Genome-wide discovery of structured noncoding RNAs in bacteria.BMC Microbiol. 2019 Mar 22;19(1):66. doi: 10.1186/s12866-019-1433-7. BMC Microbiol. 2019. PMID: 30902049 Free PMC article.
-
Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis.Nature. 2009 Dec 3;462(7273):656-9. doi: 10.1038/nature08586. Nature. 2009. PMID: 19956260 Free PMC article.
-
Small RNA gene identification and mRNA target predictions in bacteria.Bioinformatics. 2008 Dec 15;24(24):2807-13. doi: 10.1093/bioinformatics/btn560. Epub 2008 Oct 29. Bioinformatics. 2008. PMID: 18974076 Review.
Cited by
-
The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis.Nat Chem Biol. 2012 Dec;8(12):963-5. doi: 10.1038/nchembio.1095. Epub 2012 Oct 21. Nat Chem Biol. 2012. PMID: 23086297
-
Comparative genomics identifies thousands of candidate structured RNAs in human microbiomes.Genome Biol. 2021 Apr 12;22(1):100. doi: 10.1186/s13059-021-02319-w. Genome Biol. 2021. PMID: 33845850 Free PMC article.
-
Mechanisms of Evolutionary Innovation Point to Genetic Control Logic as the Key Difference Between Prokaryotes and Eukaryotes.J Mol Evol. 2015 Aug;81(1-2):34-53. doi: 10.1007/s00239-015-9688-6. Epub 2015 Jul 25. J Mol Evol. 2015. PMID: 26208881
-
Regulatory RNAs in Bacillus subtilis: a Gram-Positive Perspective on Bacterial RNA-Mediated Regulation of Gene Expression.Microbiol Mol Biol Rev. 2016 Oct 26;80(4):1029-1057. doi: 10.1128/MMBR.00026-16. Print 2016 Dec. Microbiol Mol Biol Rev. 2016. PMID: 27784798 Free PMC article. Review.
-
Small RNAs beyond Model Organisms: Have We Only Scratched the Surface?Int J Mol Sci. 2022 Apr 18;23(8):4448. doi: 10.3390/ijms23084448. Int J Mol Sci. 2022. PMID: 35457265 Free PMC article.
References
-
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

