Characterizing regulatory path motifs in integrated networks using perturbational data
- PMID: 20230615
- PMCID: PMC2864572
- DOI: 10.1186/gb-2010-11-3-r32
Characterizing regulatory path motifs in integrated networks using perturbational data
Abstract
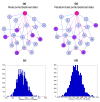
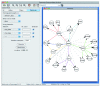
We introduce Pathicular http://bioinformatics.psb.ugent.be/software/details/Pathicular, a Cytoscape plugin for studying the cellular response to perturbations of transcription factors by integrating perturbational expression data with transcriptional, protein-protein and phosphorylation networks. Pathicular searches for 'regulatory path motifs', short paths in the integrated physical networks which occur significantly more often than expected between transcription factors and their targets in the perturbational data. A case study in Saccharomyces cerevisiae identifies eight regulatory path motifs and demonstrates their biological significance.
Figures
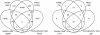

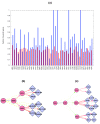
Similar articles
-
CyClus3D: a Cytoscape plugin for clustering network motifs in integrated networks.Bioinformatics. 2011 Jun 1;27(11):1587-8. doi: 10.1093/bioinformatics/btr182. Epub 2011 Apr 8. Bioinformatics. 2011. PMID: 21478195
-
Switched Latent Force Models for Reverse-Engineering Transcriptional Regulation in Gene Expression Data.IEEE/ACM Trans Comput Biol Bioinform. 2019 Jan-Feb;16(1):322-335. doi: 10.1109/TCBB.2017.2764908. Epub 2017 Oct 23. IEEE/ACM Trans Comput Biol Bioinform. 2019. PMID: 29990003
-
Integrated module and gene-specific regulatory inference implicates upstream signaling networks.PLoS Comput Biol. 2013;9(10):e1003252. doi: 10.1371/journal.pcbi.1003252. Epub 2013 Oct 17. PLoS Comput Biol. 2013. PMID: 24146602 Free PMC article.
-
The executable pathway to biological networks.Brief Funct Genomics. 2010 Jan;9(1):79-92. doi: 10.1093/bfgp/elp054. Brief Funct Genomics. 2010. PMID: 20118126 Free PMC article. Review.
-
Advances in the integration of transcriptional regulatory information into genome-scale metabolic models.Biosystems. 2016 Sep;147:1-10. doi: 10.1016/j.biosystems.2016.06.001. Epub 2016 Jun 7. Biosystems. 2016. PMID: 27287878 Review.
Cited by
-
Genome wide gene expression regulation by HIP1 Protein Interactor, HIPPI: prediction and validation.BMC Genomics. 2011 Sep 26;12:463. doi: 10.1186/1471-2164-12-463. BMC Genomics. 2011. PMID: 21943362 Free PMC article.
-
Advantages and limitations of current network inference methods.Nat Rev Microbiol. 2010 Oct;8(10):717-29. doi: 10.1038/nrmicro2419. Epub 2010 Aug 31. Nat Rev Microbiol. 2010. PMID: 20805835 Review.
-
YTRP: a repository for yeast transcriptional regulatory pathways.Database (Oxford). 2014 Mar 7;2014:bau014. doi: 10.1093/database/bau014. Print 2014. Database (Oxford). 2014. PMID: 24608172 Free PMC article.
-
Sexual Dimorphism and Aging in the Human Hyppocampus: Identification, Validation, and Impact of Differentially Expressed Genes by Factorial Microarray and Network Analysis.Front Aging Neurosci. 2016 Oct 5;8:229. doi: 10.3389/fnagi.2016.00229. eCollection 2016. Front Aging Neurosci. 2016. PMID: 27761111 Free PMC article.
-
COX2-ATP Synthase Regulates Spine Follicle Size in Hedgehogs.Int J Biol Sci. 2023 Sep 4;19(15):4763-4777. doi: 10.7150/ijbs.83387. eCollection 2023. Int J Biol Sci. 2023. PMID: 37781513 Free PMC article.
References
-
- Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

