Systematic analysis of off-target effects in an RNAi screen reveals microRNAs affecting sensitivity to TRAIL-induced apoptosis
- PMID: 20230625
- PMCID: PMC2996961
- DOI: 10.1186/1471-2164-11-175
Systematic analysis of off-target effects in an RNAi screen reveals microRNAs affecting sensitivity to TRAIL-induced apoptosis
Abstract
Background: RNA inhibition by siRNAs is a frequently used approach to identify genes required for specific biological processes. However RNAi screening using siRNAs is hampered by non-specific or off target effects of the siRNAs, making it difficult to separate genuine hits from false positives. It is thought that many of the off-target effects seen in RNAi experiments are due to siRNAs acting as microRNAs (miRNAs), causing a reduction in gene expression of unintended targets via matches to the 6 or 7 nt 'seed' sequence. We have conducted a careful examination of off-target effects during an siRNA screen for novel regulators of the TRAIL apoptosis induction pathway(s).
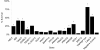
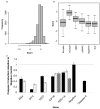
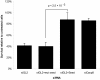
Results: We identified 3 hexamers and 3 heptamer seed sequences that appeared multiple times in the top twenty siRNAs in the TRAIL apoptosis screen. Using a novel statistical enrichment approach, we systematically identified a further 17 hexamer and 13 heptamer seed sequences enriched in high scoring siRNAs. The presence of one of these seeds sequences (which could explain 6 of 8 confirmed off-target effects) is sufficient to elicit a phenotype. Three of these seed sequences appear in the human miRNAs miR-26a, miR-145 and miR-384. Transfection of mimics of these miRNAs protects several cell types from TRAIL-induced cell death.
Conclusions: We have demonstrated a role for miR-26a, miR-145 and miR-26a in TRAIL-induced apoptosis. Further these results show that RNAi screening enriches for siRNAs with relevant off-target effects. Some of these effects can be identified by the over-representation of certain seed sequences in high-scoring siRNAs and we demonstrate the usefulness of such systematic analysis of enriched seed sequences.
Figures

Similar articles
-
Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway.Cancer Res. 2007 Nov 15;67(22):10782-8. doi: 10.1158/0008-5472.CAN-07-1484. Cancer Res. 2007. PMID: 18006822
-
Common seed analysis to identify off-target effects in siRNA screens.J Biomol Screen. 2012 Mar;17(3):370-8. doi: 10.1177/1087057111427348. Epub 2011 Nov 15. J Biomol Screen. 2012. PMID: 22086724
-
Online GESS: prediction of miRNA-like off-target effects in large-scale RNAi screen data by seed region analysis.BMC Bioinformatics. 2014 Jun 17;15:192. doi: 10.1186/1471-2105-15-192. BMC Bioinformatics. 2014. PMID: 24934636 Free PMC article.
-
Targeting microRNAs to modulate TRAIL-induced apoptosis of cancer cells.Cancer Gene Ther. 2013 Jan;20(1):33-7. doi: 10.1038/cgt.2012.81. Epub 2012 Nov 9. Cancer Gene Ther. 2013. PMID: 23138871 Review.
-
New Frontiers in Promoting TRAIL-Mediated Cell Death: Focus on Natural Sensitizers, miRNAs, and Nanotechnological Advancements.Cell Biochem Biophys. 2016 Mar;74(1):3-10. doi: 10.1007/s12013-015-0712-7. Cell Biochem Biophys. 2016. PMID: 26972296 Review.
Cited by
-
Hidden reach of the micromanagers.BMC Biol. 2010 May 11;8:53. doi: 10.1186/1741-7007-8-53. BMC Biol. 2010. PMID: 20529236 Free PMC article. Review.
-
Discovery of a dicer-independent, cell-type dependent alternate targeting sequence generator: implications in gene silencing & pooled RNAi screens.PLoS One. 2014 Jul 2;9(7):e100676. doi: 10.1371/journal.pone.0100676. eCollection 2014. PLoS One. 2014. PMID: 24987961 Free PMC article.
-
Seed-effect modeling improves the consistency of genome-wide loss-of-function screens and identifies synthetic lethal vulnerabilities in cancer cells.Genome Med. 2017 Jun 1;9(1):51. doi: 10.1186/s13073-017-0440-2. Genome Med. 2017. PMID: 28569207 Free PMC article.
-
RNA interference in Schistosoma mansoni schistosomula: selectivity, sensitivity and operation for larger-scale screening.PLoS Negl Trop Dis. 2010 Oct 19;4(10):e850. doi: 10.1371/journal.pntd.0000850. PLoS Negl Trop Dis. 2010. PMID: 20976050 Free PMC article.
-
MicroRNA-145: a potent tumour suppressor that regulates multiple cellular pathways.J Cell Mol Med. 2014 Oct;18(10):1913-26. doi: 10.1111/jcmm.12358. Epub 2014 Aug 15. J Cell Mol Med. 2014. PMID: 25124875 Free PMC article. Review.
References
-
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JCL, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nature Medicine. 1999;5(2):157–163. doi: 10.1038/5517. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

