Missing genes in the annotation of prokaryotic genomes
- PMID: 20230630
- PMCID: PMC3098052
- DOI: 10.1186/1471-2105-11-131
Missing genes in the annotation of prokaryotic genomes
Abstract
Background: Protein-coding gene detection in prokaryotic genomes is considered a much simpler problem than in intron-containing eukaryotic genomes. However there have been reports that prokaryotic gene finder programs have problems with small genes (either over-predicting or under-predicting). Therefore the question arises as to whether current genome annotations have systematically missing, small genes.
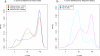
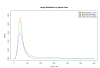
Results: We have developed a high-performance computing methodology to investigate this problem. In this methodology we compare all ORFs larger than or equal to 33 aa from all fully-sequenced prokaryotic replicons. Based on that comparison, and using conservative criteria requiring a minimum taxonomic diversity between conserved ORFs in different genomes, we have discovered 1,153 candidate genes that are missing from current genome annotations. These missing genes are similar only to each other and do not have any strong similarity to gene sequences in public databases, with the implication that these ORFs belong to missing gene families. We also uncovered 38,895 intergenic ORFs, readily identified as putative genes by similarity to currently annotated genes (we call these absent annotations). The vast majority of the missing genes found are small (less than 100 aa). A comparison of select examples with GeneMark, EasyGene and Glimmer predictions yields evidence that some of these genes are escaping detection by these programs.
Conclusions: Prokaryotic gene finders and prokaryotic genome annotations require improvement for accurate prediction of small genes. The number of missing gene families found is likely a lower bound on the actual number, due to the conservative criteria used to determine whether an ORF corresponds to a real gene.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

