A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history
- PMID: 20230632
- PMCID: PMC2907574
- DOI: 10.1186/1743-422X-7-59
A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history
Abstract
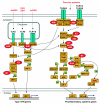
Background: Poxviruses evade the immune system of the host through the action of viral encoded inhibitors that block various signalling pathways. The exact number of viral inhibitors is not yet known. Several members of the vaccinia virus A46 and N1 families, with a Bcl-2-like structure, are involved in the regulation of the host innate immune response where they act non-redundantly at different levels of the Toll-like receptor signalling pathway. N1 also maintains an anti-apoptotic effect by acting similarly to cellular Bcl-2 proteins. Whether there are related families that could have similar functions is the main subject of this investigation.
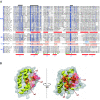
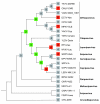
Results: We describe the sequence similarity existing among poxvirus A46, N1, N2 and C1 protein families, which share a common domain of approximately 110-140 amino acids at their C-termini that spans the entire N1 sequence. Secondary structure and fold recognition predictions suggest that this domain presents an all-alpha-helical fold compatible with the Bcl-2-like structures of vaccinia virus proteins N1, A52, B15 and K7. We propose that these protein families should be merged into a single one. We describe the phylogenetic distribution of this family and reconstruct its evolutionary history, which indicates an extensive gene gain in ancestral viruses and a further stabilization of its gene content.
Conclusions: Based on the sequence/structure similarity, we propose that other members with unknown function, like vaccinia virus N2, C1, C6 and C16/B22, might have a similar role in the suppression of host immune response as A46, A52, B15 and K7, by antagonizing at different levels with the TLR signalling pathways.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

