Site-directed spin labeling and electron paramagnetic resonance determination of vimentin head domain structure
- PMID: 20231271
- PMCID: PMC2865323
- DOI: 10.1074/jbc.M109.075598
Site-directed spin labeling and electron paramagnetic resonance determination of vimentin head domain structure
Abstract
Intermediate filament (IF) proteins have been predicted to have a conserved tripartite domain structure consisting of a largely alpha-helical central rod domain, flanked by head and tail domains. However, crystal structures have not been reported for any IF or IF protein. Although progress has been made in determining central rod domain structure, no structural data have been reported for either the head or tail domains. We used site-directed spin labeling and electron paramagnetic resonance to analyze 45 different spin labeled mutants spanning the head domain of vimentin. The data, combined with results from a previous study, provide strong evidence that the polypeptide backbones of the head domains form a symmetric dimer of closely apposed backbones that fold back onto the rod domain, imparting an asymmetry to the dimer. By following the behavior of spin labels during the process of in vitro assembly, we show that head domain structure is dynamic, changing as a result of filament assembly. Finally, because the vimentin head domain is the major site of the phosphorylation that induces disassembly at mitosis, we studied the effects of phosphorylation on head domain structure and demonstrate that phosphorylation drives specific head domain regions apart. These data provide the first evidence-based model of IF head domain structure.
Figures

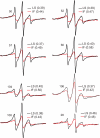
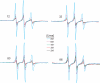
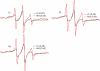
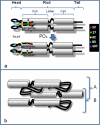
Similar articles
-
Electron paramagnetic resonance analysis of the vimentin tail domain reveals points of order in a largely disordered region and conformational adaptation upon filament assembly.Protein Sci. 2013 Jan;22(1):47-55. doi: 10.1002/pro.2182. Protein Sci. 2013. PMID: 23109052 Free PMC article.
-
Head and rod 1 interactions in vimentin: identification of contact sites, structure, and changes with phosphorylation using site-directed spin labeling and electron paramagnetic resonance.J Biol Chem. 2009 Mar 13;284(11):7330-8. doi: 10.1074/jbc.M809029200. Epub 2008 Dec 31. J Biol Chem. 2009. PMID: 19117942 Free PMC article.
-
Structural characterization of human vimentin rod 1 and the sequencing of assembly steps in intermediate filament formation in vitro using site-directed spin labeling and electron paramagnetic resonance.J Biol Chem. 2004 Oct 22;279(43):44841-6. doi: 10.1074/jbc.M406257200. Epub 2004 Jul 1. J Biol Chem. 2004. PMID: 15231822 Free PMC article.
-
Describing the structure and assembly of protein filaments by EPR spectroscopy of spin-labeled side chains.Cell Biochem Biophys. 2007;48(1):45-53. doi: 10.1007/s12013-007-0035-4. Cell Biochem Biophys. 2007. PMID: 17703067 Review.
-
Elucidating the design principles of photosynthetic electron-transfer proteins by site-directed spin labeling EPR spectroscopy.Biochim Biophys Acta. 2016 May;1857(5):548-556. doi: 10.1016/j.bbabio.2015.08.009. Epub 2015 Sep 1. Biochim Biophys Acta. 2016. PMID: 26334844 Review.
Cited by
-
Use of electron paramagnetic resonance to solve biochemical problems.Biochemistry. 2013 Sep 3;52(35):5967-84. doi: 10.1021/bi400834a. Epub 2013 Aug 20. Biochemistry. 2013. PMID: 23961941 Free PMC article. Review.
-
Molecular structure of soluble vimentin tetramers.Sci Rep. 2023 May 31;13(1):8841. doi: 10.1038/s41598-023-34814-4. Sci Rep. 2023. PMID: 37258554 Free PMC article.
-
Vimentin in cancer and its potential as a molecular target for cancer therapy.Cell Mol Life Sci. 2011 Sep;68(18):3033-46. doi: 10.1007/s00018-011-0735-1. Epub 2011 Jun 3. Cell Mol Life Sci. 2011. PMID: 21637948 Free PMC article. Review.
-
Electron paramagnetic resonance analysis of the vimentin tail domain reveals points of order in a largely disordered region and conformational adaptation upon filament assembly.Protein Sci. 2013 Jan;22(1):47-55. doi: 10.1002/pro.2182. Protein Sci. 2013. PMID: 23109052 Free PMC article.
-
Phosphoproteome of Toxoplasma gondii Infected Host Cells Reveals Specific Cellular Processes Predominating in Different Phases of Infection.Am J Trop Med Hyg. 2017 Jul;97(1):236-244. doi: 10.4269/ajtmh.16-0901. Am J Trop Med Hyg. 2017. PMID: 28719319 Free PMC article.
References
-
- Hanukoglu I., Fuchs E. (1983) Cell 33, 915–924 - PubMed
-
- Steinert P. M., Parry D. A. (1985) Annu. Rev. Cell Biol. 1, 41–65 - PubMed
-
- Albers K., Fuchs E. (1992) Int. Rev. Cytol. 134, 243–279 - PubMed
-
- Fuchs E., Hanukoglu I. (1983) Cell 34, 332–334 - PubMed
-
- Herrmann H., Aebi U. (2004) Annu. Rev. Biochem. 73, 749–789 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

