Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts
- PMID: 20231276
- PMCID: PMC2863236
- DOI: 10.1074/jbc.M110.115345
Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts
Abstract
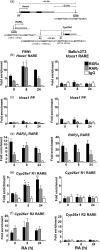
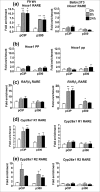
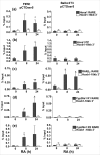
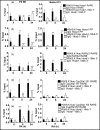
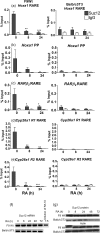
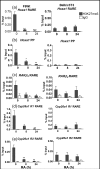
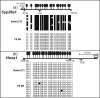
Retinoic acid (RA), a vitamin A metabolite, regulates transcription by binding to RA receptor (RAR) and retinoid X receptor (RXR) heterodimers. This transcriptional response is determined by receptor interactions with transcriptional regulators and chromatin modifying proteins. We compared transcriptional responses of three RA target genes (Hoxa1, Cyp26a1, RARbeta(2)) in primary embryo fibroblasts (mouse embryonic fibroblasts), immortalized fibroblasts (Balb/c3T3), and F9 teratocarcinoma stem cells. Hoxa1 and Cyp26a1 transcripts are not expressed, but RARbeta(2) transcripts are induced by RA in mouse embryonic fibroblasts and Balb/c3T3 cells. Retinoid receptors (RARgamma, RXRalpha), coactivators (pCIP (NCOA3, SRC3)), and p300 and RNA polymerase II are recruited only to the RARbeta(2) RA response element (RARE) in Balb/c3T3, whereas these proteins are recruited to RAREs of all three genes by RA in F9 cells. In F9, RA reduces polycomb (PcG) protein Suz12 and the associated H3K27me3 repressive epigenetic modification at the RAREs of all three genes. In contrast, in Balb/c3T3 cells cultured in the +/-RA, Suz12 is not associated with the Hoxa1, RARbeta(2), and Cyp26a1 RAREs, whereas slow levels of the H3K27me3 mark are seen at these RAREs. Thus, Suz12 is not required for gene repression in the absence of RA. Even though the Hoxa1 RARE and proximal promoter show high levels of H3K9,K14 acetylation in Balb/c3T3, the Hoxa1 gene is not transcriptionally activated by RA. In Balb/c3T3, CpG islands are methylated in the Cyp26a1 promoter region but not in the Hoxa1 promoter or in these promoters in F9 cells. We have delineated the complex mechanisms that control RA-mediated transcription in fibroblasts versus stem cells.
Figures


Similar articles
-
Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells.J Mol Biol. 2007 Sep 14;372(2):298-316. doi: 10.1016/j.jmb.2007.06.079. Epub 2007 Jul 3. J Mol Biol. 2007. PMID: 17663992 Free PMC article.
-
Retinoic acid and histone deacetylases regulate epigenetic changes in embryonic stem cells.J Biol Chem. 2014 Jul 11;289(28):19519-30. doi: 10.1074/jbc.M114.556555. Epub 2014 May 12. J Biol Chem. 2014. PMID: 24821725 Free PMC article.
-
Retinoic acid receptor isotype specificity in F9 teratocarcinoma stem cells results from the differential recruitment of coregulators to retinoic response elements.J Biol Chem. 2007 Nov 16;282(46):33421-33434. doi: 10.1074/jbc.M704845200. Epub 2007 Sep 17. J Biol Chem. 2007. PMID: 17875646
-
Cytochrome P450s in the regulation of cellular retinoic acid metabolism.Annu Rev Nutr. 2011 Aug 21;31:65-87. doi: 10.1146/annurev-nutr-072610-145127. Annu Rev Nutr. 2011. PMID: 21529158 Free PMC article. Review.
-
Retinoids induce stem cell differentiation via epigenetic changes.Semin Cell Dev Biol. 2013 Dec;24(10-12):701-5. doi: 10.1016/j.semcdb.2013.08.002. Epub 2013 Aug 21. Semin Cell Dev Biol. 2013. PMID: 23973942 Free PMC article. Review.
Cited by
-
Discovery of retinoic acid receptor agonists as proliferators of cardiac progenitor cells through a phenotypic screening approach.Stem Cells Transl Med. 2020 Jan;9(1):47-60. doi: 10.1002/sctm.19-0069. Epub 2019 Sep 11. Stem Cells Transl Med. 2020. PMID: 31508905 Free PMC article.
-
Increasing adipocyte number and reducing adipocyte size: the role of retinoids in adipose tissue development and metabolism.Crit Rev Food Sci Nutr. 2024;64(29):10608-10625. doi: 10.1080/10408398.2023.2227258. Epub 2023 Jul 10. Crit Rev Food Sci Nutr. 2024. PMID: 37427553 Free PMC article. Review.
-
Retinoids regulate stem cell differentiation.J Cell Physiol. 2011 Feb;226(2):322-30. doi: 10.1002/jcp.22417. J Cell Physiol. 2011. PMID: 20836077 Free PMC article. Review.
-
Targeting the epigenome with bioactive food components for cancer prevention.J Nutrigenet Nutrigenomics. 2011;4(5):275-92. doi: 10.1159/000334585. Epub 2012 Feb 22. J Nutrigenet Nutrigenomics. 2011. PMID: 22353664 Free PMC article. Review.
-
Retinoic acid drives surface epithelium fate determination through the TCF7-MSX2 axis.Cell Mol Life Sci. 2024 Dec 27;82(1):16. doi: 10.1007/s00018-024-05525-4. Cell Mol Life Sci. 2024. PMID: 39725818 Free PMC article.
References
-
- Niederreither K., Dollé P. (2008) Nat. Rev. Genet. 9, 541–553 - PubMed
-
- Gudas L. J. (1994) J. Biol. Chem. 269, 15399–15402 - PubMed
-
- Blomhoff R., Blomhoff H. K. (2006) J. Neurobiol. 66, 606–630 - PubMed
-
- Bastien J., Rochette-Egly C. (2004) Gene 328, 1–16 - PubMed
-
- Kastner P., Mark M., Ghyselinck N., Krezel W., Dupé V., Grondona J. M., Chambon P. (1997) Development 124, 313–326 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

