CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast
- PMID: 20231317
- PMCID: PMC2841337
- DOI: 10.1101/gad.1883410
CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast
Abstract
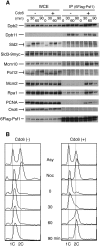
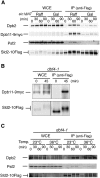
Eukaryotic chromosomal DNA replication requires cyclin-dependent kinase (CDK) activity. CDK phosphorylates two yeast replication proteins, Sld3 and Sld2, both of which bind to Dpb11 when phosphorylated. These phosphorylation-dependent interactions are essential and are the minimal requirements for CDK-dependent activation of DNA replication. However, how these interactions activate DNA replication has not been elucidated. Here, we show that CDK promotes the formation of a newly identified fragile complex, the preloading complex (pre-LC) containing DNA polymerase epsilon (Pol epsilon), GINS, Sld2, and Dpb11. Formation of the pre-LC requires phosphorylation of Sld2 by CDK, but is independent of DNA replication, protein association with replication origins, and Dbf4-dependent Cdc7 kinase, which is also essential for the activation of DNA replication. We also demonstrate that Pol epsilon, GINS, Dpb11, and CDK-phosphorylated Sld2 form a complex in vitro. The genetic interactions between Pol epsilon, GINS, Sld2, and Dpb11 suggest further that they form an essential complex in cells. We propose that CDK regulates the initiation of DNA replication in budding yeast through formation of the pre-LC.
Figures


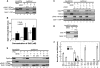


References
-
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase ɛ by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. - PubMed
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. - PubMed
-
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ɛ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
