Forensic identification using skin bacterial communities
- PMID: 20231444
- PMCID: PMC2852011
- DOI: 10.1073/pnas.1000162107
Forensic identification using skin bacterial communities
Abstract
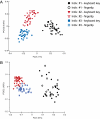
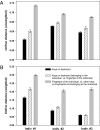
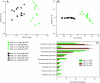
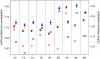
Recent work has demonstrated that the diversity of skin-associated bacterial communities is far higher than previously recognized, with a high degree of interindividual variability in the composition of bacterial communities. Given that skin bacterial communities are personalized, we hypothesized that we could use the residual skin bacteria left on objects for forensic identification, matching the bacteria on the object to the skin-associated bacteria of the individual who touched the object. Here we describe a series of studies de-monstrating the validity of this approach. We show that skin-associated bacteria can be readily recovered from surfaces (including single computer keys and computer mice) and that the structure of these communities can be used to differentiate objects handled by different individuals, even if those objects have been left untouched for up to 2 weeks at room temperature. Furthermore, we demonstrate that we can use a high-throughput pyrosequencing-based ap-proach to quantitatively compare the bacterial communities on objects and skin to match the object to the individual with a high degree of certainty. Although additional work is needed to further establish the utility of this approach, this series of studies introduces a forensics approach that could eventually be used to independently evaluate results obtained using more traditional forensic practices.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Harnessing the power of the human microbiome.Proc Natl Acad Sci U S A. 2010 Apr 6;107(14):6125-6. doi: 10.1073/pnas.1002112107. Epub 2010 Apr 1. Proc Natl Acad Sci U S A. 2010. PMID: 20360554 Free PMC article. No abstract available.
Similar articles
-
Microbial DNA fingerprinting of human fingerprints: dynamic colonization of fingertip microflora challenges human host inferences for forensic purposes.Int J Legal Med. 2010 Sep;124(5):477-81. doi: 10.1007/s00414-009-0352-9. Epub 2009 Jun 24. Int J Legal Med. 2010. PMID: 19551400 Free PMC article.
-
The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations.PLoS One. 2015 Oct 28;10(10):e0141842. doi: 10.1371/journal.pone.0141842. eCollection 2015. PLoS One. 2015. PMID: 26510185 Free PMC article.
-
Harnessing the power of the human microbiome.Proc Natl Acad Sci U S A. 2010 Apr 6;107(14):6125-6. doi: 10.1073/pnas.1002112107. Epub 2010 Apr 1. Proc Natl Acad Sci U S A. 2010. PMID: 20360554 Free PMC article. No abstract available.
-
Skin Microbiome Analysis for Forensic Human Identification: What Do We Know So Far?Microorganisms. 2020 Jun 9;8(6):873. doi: 10.3390/microorganisms8060873. Microorganisms. 2020. PMID: 32527009 Free PMC article. Review.
-
The airway microbiome in cystic fibrosis and implications for treatment.Curr Opin Pediatr. 2011 Jun;23(3):319-24. doi: 10.1097/MOP.0b013e32834604f2. Curr Opin Pediatr. 2011. PMID: 21494150 Review.
Cited by
-
Tracking Strains in the Microbiome: Insights from Metagenomics and Models.Front Microbiol. 2016 May 19;7:712. doi: 10.3389/fmicb.2016.00712. eCollection 2016. Front Microbiol. 2016. PMID: 27242733 Free PMC article. Review.
-
Recovery of microbial DNA by agar-containing solution from extremely low-biomass specimens including skin.Sci Rep. 2023 Nov 11;13(1):19666. doi: 10.1038/s41598-023-46890-7. Sci Rep. 2023. PMID: 37952000 Free PMC article.
-
Pyrosequencing as a tool for better understanding of human microbiomes.J Oral Microbiol. 2012;4. doi: 10.3402/jom.v4i0.10743. Epub 2012 Jan 23. J Oral Microbiol. 2012. PMID: 22279602 Free PMC article.
-
The Human Microbiome Project: a community resource for the healthy human microbiome.PLoS Biol. 2012;10(8):e1001377. doi: 10.1371/journal.pbio.1001377. Epub 2012 Aug 14. PLoS Biol. 2012. PMID: 22904687 Free PMC article.
-
Challenges in Human Skin Microbial Profiling for Forensic Science: A Review.Genes (Basel). 2020 Aug 28;11(9):1015. doi: 10.3390/genes11091015. Genes (Basel). 2020. PMID: 32872386 Free PMC article. Review.
References
-
- Jarvis WR. Handwashing—the Semmelweis lesson forgotten? Lancet. 1994;344:1311–1312. - PubMed
-
- Pittet D, Allegranzi B, Boyce J, World Health Organization World Alliance for Patient Safety First Global Patient Safety Challenge Core Group of Experts The World Health Organization guidelines on hand hygiene in health care and their consensus recommendations. Infect Control Hosp Epidemiol. 2009;30:611–622. - PubMed
-
- Smith SM, Eng RHK, Padberg FT., Jr Survival of nosocomial pathogenic bacteria at ambient temperature. J Med. 1996;27:293–302. - PubMed
-
- Brooke JS, Annand JW, Hammer A, Dembkowski K, Shulman ST. Investigation of bacterial pathogens on 70 frequently used environmental surfaces in a large urban U.S. university. J Environ Health. 2009;71:17–22. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

