Ordering human CD34+CD10-CD19+ pre/pro-B-cell and CD19- common lymphoid progenitor stages in two pro-B-cell development pathways
- PMID: 20231472
- PMCID: PMC2851857
- DOI: 10.1073/pnas.0907942107
Ordering human CD34+CD10-CD19+ pre/pro-B-cell and CD19- common lymphoid progenitor stages in two pro-B-cell development pathways
Abstract
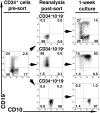
Studies here respond to two long-standing questions: Are human "pre/pro-B" CD34(+)CD10(-)CD19(+) and "common lymphoid progenitor (CLP)/early-B" CD34(+)CD10(+)CD19(-) alternate precursors to "pro-B" CD34(+)CD19(+)CD10(+) cells, and do the pro-B cells that arise from these progenitors belong to the same or distinct B-cell development pathways? Using flow cytometry, gene expression profiling, and Ig V(H)-D-J(H) sequencing, we monitor the initial 10 generations of development of sorted cord blood CD34(high)Lineage(-) pluripotential progenitors growing in bone marrow S17 stroma cocultures. We show that (i) multipotent progenitors (CD34(+)CD45RA(+)CD10(-)CD19(-)) directly generate an initial wave of Pax5(+)TdT(-) "unilineage" pre/pro-B cells and a later wave of "multilineage" CLP/early-B cells and (ii) the cells generated in these successive stages act as precursors for distinct pro-B cells through two independent layered pathways. Studies by others have tracked the origin of B-lineage leukemias in elderly mice to the mouse B-1a pre/pro-B lineage, which lacks the TdT activity that diversifies the V(H)-D-J(H) Ig heavy chain joints found in the early-B or B-2 lineage. Here, we show a similar divergence in human B-cell development pathways between the Pax5(+)TdT(-) pre/pro-B differentiation pathway that gives rise to infant B-lineage leukemias and the early-B pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Human bone marrow stromal cells simultaneously support B and T/NK lineage development from human haematopoietic progenitors: a principal role for flt3 ligand in lymphopoiesis.Br J Haematol. 2012 Jun;157(6):674-86. doi: 10.1111/j.1365-2141.2012.09109.x. Epub 2012 Mar 30. Br J Haematol. 2012. PMID: 22463758 Clinical Trial.
-
Characterization of CD7+CD19+ lymphoid cells after Epstein-Barr virus transformation.J Immunol. 1993 Jul 1;151(1):92-9. J Immunol. 1993. PMID: 7686950
-
The growth response to IL-7 during normal human B cell ontogeny is restricted to B-lineage cells expressing CD34.J Immunol. 1995 Jan 1;154(1):58-67. J Immunol. 1995. PMID: 7527823
-
Bone marrow microenvironmental changes in aged mice compromise V(D)J recombinase activity and B cell generation.Semin Immunol. 2005 Oct;17(5):347-55. doi: 10.1016/j.smim.2005.05.012. Semin Immunol. 2005. PMID: 15963731 Review.
-
B-lymphocyte commitment: identifying the point of no return.Semin Immunol. 2011 Oct;23(5):335-40. doi: 10.1016/j.smim.2011.08.005. Epub 2011 Sep 23. Semin Immunol. 2011. PMID: 21944938 Review.
Cited by
-
Differential expression of the transcription factor ARID3a in lupus patient hematopoietic progenitor cells.J Immunol. 2015 Feb 1;194(3):940-9. doi: 10.4049/jimmunol.1401941. Epub 2014 Dec 22. J Immunol. 2015. PMID: 25535283 Free PMC article.
-
ARID3a expression in human hematopoietic stem cells is associated with distinct gene patterns in aged individuals.Immun Ageing. 2020 Sep 3;17:24. doi: 10.1186/s12979-020-00198-6. eCollection 2020. Immun Ageing. 2020. PMID: 32905435 Free PMC article.
-
Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development.Cell. 2014 Apr 24;157(3):714-25. doi: 10.1016/j.cell.2014.04.005. Cell. 2014. PMID: 24766814 Free PMC article.
-
Stemness of B-cell progenitors in multiple myeloma bone marrow.Clin Cancer Res. 2012 Nov 15;18(22):6155-68. doi: 10.1158/1078-0432.CCR-12-0531. Epub 2012 Sep 17. Clin Cancer Res. 2012. PMID: 22988056 Free PMC article.
-
An optimized workflow for single-cell transcriptomics and repertoire profiling of purified lymphocytes from clinical samples.Sci Rep. 2020 Feb 10;10(1):2219. doi: 10.1038/s41598-020-58939-y. Sci Rep. 2020. PMID: 32042039 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

