Whole genome survey of copy number variation in the spontaneously hypertensive rat: relationship to quantitative trait loci, gene expression, and blood pressure
- PMID: 20231529
- PMCID: PMC5266550
- DOI: 10.1161/HYPERTENSIONAHA.109.141663
Whole genome survey of copy number variation in the spontaneously hypertensive rat: relationship to quantitative trait loci, gene expression, and blood pressure
Erratum in
- Hypertension. 2010 Jun;55(6):e28
Abstract
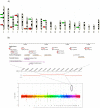
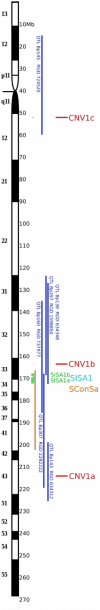
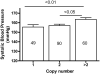
Copy number variation has emerged recently as an important genetic mechanism leading to phenotypic heterogeneity. The aim of our study was to determine whether copy number variants (CNVs) exist between the spontaneously hypertensive rat (SHR) and its control strain, the Wistar-Kyoto rat, whether these map to quantitative trait loci in the rat and whether CNVs associate with gene expression or blood pressure differences between the 2 strains. We performed a comparative genomic hybridization assay between SHR and Wistar-Kyoto strains using a whole-genome array. In total, 16 CNVs were identified and validated (6 because of a relative loss of copy number in the SHR and 10 because of a relative gain). CNVs were present on rat autosomes 1, 3, 4, 6, 7, 10, 14, and 17 and varied in size from 10.0 kb to 1.6 Mb. Most of these CNVs mapped to chromosomal regions within previously identified quantitative trait loci, including those for blood pressure in the SHR. Transcriptomic experiments confirmed differences in the renal expression of several genes (including Ms4a6a, Ndrg3, Egln1, Cd36, Sema3a, Ugt2b, and Idi21) located in some of the CNVs between SHR and Wistar-Kyoto rats. In F(2) animals derived from an SHRxWistar-Kyoto cross, we also found a significant increase in blood pressure associated with an increase in copy number in the Egln1 gene. Our findings suggest that CNVs may play a role in the susceptibility to hypertension and related traits in the SHR.
Figures
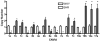
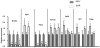
Similar articles
-
Combined genealogical, mapping, and expression approaches to identify spontaneously hypertensive rat hypertension candidate genes.Hypertension. 2005 Apr;45(4):698-704. doi: 10.1161/01.HYP.0000156498.78896.37. Epub 2005 Feb 14. Hypertension. 2005. PMID: 15710778
-
Dissection of chromosome 18 blood pressure and salt-sensitivity quantitative trait loci in the spontaneously hypertensive rat.Hypertension. 2009 Sep;54(3):639-45. doi: 10.1161/HYPERTENSIONAHA.108.126664. Epub 2009 Jul 20. Hypertension. 2009. PMID: 19620519 Free PMC article.
-
Genetic, physiological and comparative genomic studies of hypertension and insulin resistance in the spontaneously hypertensive rat.Dis Model Mech. 2017 Mar 1;10(3):297-306. doi: 10.1242/dmm.026716. Epub 2017 Jan 26. Dis Model Mech. 2017. PMID: 28130354 Free PMC article.
-
Recent advances in genetics of the spontaneously hypertensive rat.Curr Hypertens Rep. 2010 Feb;12(1):5-9. doi: 10.1007/s11906-009-0083-9. Curr Hypertens Rep. 2010. PMID: 20425152 Free PMC article. Review.
-
[Cosegregation studies in spontaneously hypertensive rats].Nihon Rinsho. 1993 Jun;51(6):1602-9. Nihon Rinsho. 1993. PMID: 8320840 Review. Japanese.
Cited by
-
Genome-wide detection of copy number variations among diverse horse breeds by array CGH.PLoS One. 2014 Jan 30;9(1):e86860. doi: 10.1371/journal.pone.0086860. eCollection 2014. PLoS One. 2014. PMID: 24497987 Free PMC article.
-
Whole rat DNA array survey for candidate genes related to hypertension in kidneys from three spontaneously hypertensive rat substrains at two stages of age and with hypotensive induction caused by hydralazine hydrochloride.Exp Ther Med. 2011 Mar;2(2):201-212. doi: 10.3892/etm.2011.193. Epub 2011 Jan 14. Exp Ther Med. 2011. PMID: 22977489 Free PMC article.
-
Genomic Copy Number Variation Study of Nine Macaca Species Provides New Insights into Their Genetic Divergence, Adaptation, and Biomedical Application.Genome Biol Evol. 2020 Dec 6;12(12):2211-2230. doi: 10.1093/gbe/evaa200. Genome Biol Evol. 2020. PMID: 32970804 Free PMC article.
-
Integrated genomic approaches to identification of candidate genes underlying metabolic and cardiovascular phenotypes in the spontaneously hypertensive rat.Physiol Genomics. 2011 Nov 7;43(21):1207-18. doi: 10.1152/physiolgenomics.00210.2010. Epub 2011 Aug 16. Physiol Genomics. 2011. PMID: 21846806 Free PMC article.
-
Array-comparative genomic hybridization reveals loss of SOCS6 is associated with poor prognosis in primary lung squamous cell carcinoma.PLoS One. 2012;7(2):e30398. doi: 10.1371/journal.pone.0030398. Epub 2012 Feb 17. PLoS One. 2012. PMID: 22363434 Free PMC article.
References
-
- Charchar FJ, Zimmerli LU, Tomaszewski M. The pressure of finding human hypertension genes: new tools, old dilemmas. J Hum Hypertens. 2008;22:821–828. - PubMed
-
- Okamoto K, Aoki K. Development of a strain of spontaneously hypertensive rats. Jpn Circ J. 1963;27:282–293. - PubMed
-
- Pravenec M, Kren V. Genetic analysis of complex cardiovascular traits in the spontaneously hypertensive rat. Exp Physiol. 2005;90:273–276. - PubMed
-
- Cowley AW., Jr. The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–840. - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito N, Healy J, Hicks J, Ye K, Reiner A, Gilliam CC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Sci. 2004;305:525–528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

