Transcriptional regulation of Rex1 (zfp42) in normal prostate epithelial cells and prostate cancer cells
- PMID: 20232320
- PMCID: PMC3306262
- DOI: 10.1002/jcp.22071
Transcriptional regulation of Rex1 (zfp42) in normal prostate epithelial cells and prostate cancer cells
Abstract
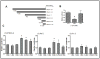
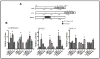
Rex1 (zfp42) was identified by our laboratory because of its reduced expression in F9 teratocarcinoma stem cells after retinoic acid (RA) treatment. The Rex1 (Zfp42) gene is currently widely used as a marker of embryonic stem cells. We compared the transcriptional regulation of the human Rex1 gene in NTera-2 (NT-2) human teratocarcinoma, normal human prostate epithelial cells (PrEC), and prostate cancer cells (PC-3) by promoter/luciferase analyses. Oct4, Sox2, Nanog, and Dax1 transcripts are expressed at higher levels in NT-2 and PrEC cells than in PC-3 cells. Co-transfection analyses showed that YY1 and Rex1 are positive regulators of hRex1 transcription in NT-2 and PrEC cells, whereas Nanog is not. Serial deletion constructs of the hRex1 promoter were created and analyzed, by which we identified a potential negative regulatory site that is located between -1 and -0.4 kb of the hRex1 promoter. We also delineated regions of the hRex1 promoter between -0.4 kb and the TSS that, when mutated, reduced transcriptional activation; these are putative Rex1 binding sites. Mutation of a putative Rex1 binding site in electrophoretic mobility shift assays (EMSA) resulted in reduced protein binding. Taken together, our results indicate that hRex1 binds to the hRex1 promoter region at -298 bp and positively regulates hRex1 transcription, but that this regulation is lost in PC-3 human prostate cancer cells. This lack of positive transcriptional regulation by the hRex1 protein may be responsible for the lack of Rex1 expression in PC-3 prostate cancer cells.
(c) 2010 Wiley-Liss, Inc.
Figures




Similar articles
-
Transcriptional activation of the suppressor of cytokine signaling-3 (SOCS-3) gene via STAT3 is increased in F9 REX1 (ZFP-42) knockout teratocarcinoma stem cells relative to wild-type cells.J Mol Biol. 2008 Mar 14;377(1):28-46. doi: 10.1016/j.jmb.2007.12.038. Epub 2008 Jan 30. J Mol Biol. 2008. PMID: 18237746 Free PMC article.
-
Retinoic acid receptors and GATA transcription factors activate the transcription of the human lecithin:retinol acyltransferase gene.Int J Biochem Cell Biol. 2009 Mar;41(3):546-53. doi: 10.1016/j.biocel.2008.06.007. Epub 2008 Jul 4. Int J Biochem Cell Biol. 2009. PMID: 18652909 Free PMC article.
-
The putative human stem cell marker, Rex-1 (Zfp42): structural classification and expression in normal human epithelial and carcinoma cell cultures.Mol Carcinog. 2006 Dec;45(12):887-900. doi: 10.1002/mc.20186. Mol Carcinog. 2006. PMID: 16865673
-
Pluripotent genes in avian stem cells.Dev Growth Differ. 2013 Jan;55(1):41-51. doi: 10.1111/dgd.12021. Epub 2012 Dec 20. Dev Growth Differ. 2013. PMID: 23278808 Review.
-
Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication.J Exp Clin Cancer Res. 2016 May 25;35:84. doi: 10.1186/s13046-016-0359-2. J Exp Clin Cancer Res. 2016. PMID: 27225481 Free PMC article. Review.
Cited by
-
Mechanisms Regulating Stemness and Differentiation in Embryonal Carcinoma Cells.Stem Cells Int. 2017;2017:3684178. doi: 10.1155/2017/3684178. Epub 2017 Mar 8. Stem Cells Int. 2017. PMID: 28373885 Free PMC article. Review.
-
A ZFP42/MARK2 regulatory network reduces the damage of retinal ganglion cells in glaucoma: a study based on GEO dataset and in vitro experiments.Apoptosis. 2022 Dec;27(11-12):1049-1059. doi: 10.1007/s10495-022-01746-9. Epub 2022 Sep 21. Apoptosis. 2022. PMID: 36131186
-
REX1 promotes EMT-induced cell metastasis by activating the JAK2/STAT3-signaling pathway by targeting SOCS1 in cervical cancer.Oncogene. 2019 Oct;38(43):6940-6957. doi: 10.1038/s41388-019-0906-3. Epub 2019 Aug 13. Oncogene. 2019. PMID: 31409905
-
Embryonic stem cell markers.Molecules. 2012 May 25;17(6):6196-236. doi: 10.3390/molecules17066196. Molecules. 2012. PMID: 22634835 Free PMC article. Review.
-
The Role of Nuclear Receptors in Prostate Cancer.Cells. 2019 Jun 17;8(6):602. doi: 10.3390/cells8060602. Cells. 2019. PMID: 31212954 Free PMC article. Review.
References
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. - PubMed
-
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. - PubMed
-
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol 2009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

