Improving binding specificity of pharmacological chaperones that target mutant superoxide dismutase-1 linked to familial amyotrophic lateral sclerosis using computational methods
- PMID: 20232802
- PMCID: PMC2881568
- DOI: 10.1021/jm901062p
Improving binding specificity of pharmacological chaperones that target mutant superoxide dismutase-1 linked to familial amyotrophic lateral sclerosis using computational methods
Abstract
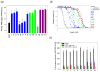
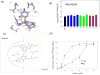
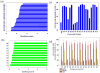
We recently described a set of drug-like molecules obtained from an in silico screen that stabilize mutant superoxide dismutase-1 (SOD-1) linked to familial amyotrophic lateral sclerosis (ALS) against unfolding and aggregation but exhibited poor binding specificity toward SOD-1 in presence of blood plasma. A reasonable but not a conclusive model for the binding of these molecules was proposed on the basis of restricted docking calculations and site-directed mutagenesis of key residues at the dimer interface. A set of hydrogen bonding constraints obtained from these experiments were used to guide docking calculations with compound library around the dimer interface. A series of chemically unrelated hits were predicted, which were experimentally tested for their ability to block aggregation. At least six of the new molecules exhibited high specificity of binding toward SOD-1 in the presence of blood plasma. These molecules represent a new class of molecules for further development into clinical candidates.
Figures

References
-
- Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. - PubMed
-
- Dobson CM. Protein aggregation and its consequences for human disease. Protein Pept Lett. 2006;13:219–227. - PubMed
-
- Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. - PubMed
-
- Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des. 1998;3:R9–23. - PubMed
-
- Fowler DM, Kelly JW. Aggregating knowledge about prions and amyloid. Cell. 2009;137:20–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

