DNA architecture, deformability, and nucleosome positioning
- PMID: 20232929
- PMCID: PMC3000044
- DOI: 10.1080/073911010010524943
DNA architecture, deformability, and nucleosome positioning
Abstract
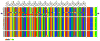
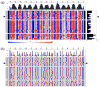
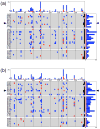
The positioning of DNA on nucleosomes is critical to both the organization and expression of the genetic message. Here we focus on DNA conformational signals found in the growing library of known high-resolution core-particle structures and the ways in which these features may contribute to the positioning of nucleosomes on specific DNA sequences. We survey the chemical composition of the protein-DNA assemblies and extract features along the DNA superhelical pathway - the minor-groove width and the deformations of successive base pairs - determined with reasonable accuracy in the structures. We also examine the extent to which the various nucleosome core-particle structures accommodate the observed settings of the crystallized sequences and the known positioning of the high-affinity synthetic '601' sequence on DNA. We 'thread' these sequences on the different structural templates and estimate the cost of each setting with knowledge-based potentials that reflect the conformational properties of the DNA base-pair steps in other high-resolution protein-bound complexes.
Figures


Similar articles
-
Structure-based analysis of DNA sequence patterns guiding nucleosome positioning in vitro.J Biomol Struct Dyn. 2010 Jun;27(6):821-41. doi: 10.1080/073911010010524947. J Biomol Struct Dyn. 2010. PMID: 20232936 Free PMC article.
-
The structure of DNA in the nucleosome core.Nature. 2003 May 8;423(6936):145-50. doi: 10.1038/nature01595. Nature. 2003. PMID: 12736678
-
DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences.Biophys J. 2009 Mar 18;96(6):2245-60. doi: 10.1016/j.bpj.2008.11.040. Biophys J. 2009. PMID: 19289051 Free PMC article.
-
Working the kinks out of nucleosomal DNA.Curr Opin Struct Biol. 2011 Jun;21(3):348-57. doi: 10.1016/j.sbi.2011.03.006. Epub 2011 Apr 7. Curr Opin Struct Biol. 2011. PMID: 21482100 Free PMC article. Review.
-
A structural perspective on the where, how, why, and what of nucleosome positioning.J Biomol Struct Dyn. 2010 Jun;27(6):803-20. doi: 10.1080/07391102.2010.10508585. J Biomol Struct Dyn. 2010. PMID: 20232935 Review.
Cited by
-
The intrinsic mechanics of B-DNA in solution characterized by NMR.Nucleic Acids Res. 2016 Apr 20;44(7):3432-47. doi: 10.1093/nar/gkw084. Epub 2016 Feb 15. Nucleic Acids Res. 2016. PMID: 26883628 Free PMC article.
-
Quantitative contribution of the spacer length in the supercoiling-sensitivity of bacterial promoters.Nucleic Acids Res. 2022 Jul 22;50(13):7287-7297. doi: 10.1093/nar/gkac579. Nucleic Acids Res. 2022. PMID: 35776118 Free PMC article.
-
High-resolution biophysical analysis of the dynamics of nucleosome formation.Sci Rep. 2016 Jun 6;6:27337. doi: 10.1038/srep27337. Sci Rep. 2016. PMID: 27263658 Free PMC article.
-
Insights into Gene Expression and Packaging from Computer Simulations.Biophys Rev. 2012 Sep 1;4(3):171-178. doi: 10.1007/s12551-012-0093-8. Biophys Rev. 2012. PMID: 23139731 Free PMC article.
-
Physical properties of naked DNA influence nucleosome positioning and correlate with transcription start and termination sites in yeast.BMC Genomics. 2011 Oct 7;12:489. doi: 10.1186/1471-2164-12-489. BMC Genomics. 2011. PMID: 21981773 Free PMC article.
References
-
- Kornberg RD, Lorch Y. Cell. 1999;98:285–294. - PubMed
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. - PubMed
-
- Harp JM, Hanson BL, Timm DE, Bunick GJ. Acta Crystallographica Section D - Biological Crystallography. 2000;56:1513–1534. - PubMed
-
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Nature Structural Biology. 2000;7:1121–1124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
