MapMi: automated mapping of microRNA loci
- PMID: 20233390
- PMCID: PMC2858034
- DOI: 10.1186/1471-2105-11-133
MapMi: automated mapping of microRNA loci
Abstract
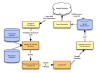
Background: A large effort to discover microRNAs (miRNAs) has been under way. Currently miRBase is their primary repository, providing annotations of primary sequences, precursors and probable genomic loci. In many cases miRNAs are identical or very similar between related (or in some cases more distant) species. However, miRBase focuses on those species for which miRNAs have been directly confirmed. Secondly, specific miRNAs or their loci are sometimes not annotated even in well-covered species. We sought to address this problem by developing a computational system for automated mapping of miRNAs within and across species. Given the sequence of a known miRNA in one species it is relatively straightforward to determine likely loci of that miRNA in other species. Our primary goal is not the discovery of novel miRNAs but the mapping of validated miRNAs in one species to their most likely orthologues in other species.
Results: We present MapMi, a computational system for automated miRNA mapping across and within species. This method has a sensitivity of 92.20% and a specificity of 97.73%. Using the latest release (v14) of miRBase, we obtained 10,944 unannotated potential miRNAs when MapMi was applied to all 21 species in Ensembl Metazoa release 2 and 46 species from Ensembl release 55.
Conclusions: The pipeline and an associated web-server for mapping miRNAs are freely available on http://www.ebi.ac.uk/enright-srv/MapMi/. In addition precomputed miRNA mappings of miRBase miRNAs across a large number of species are provided.
Figures

Similar articles
-
miRBase: the microRNA sequence database.Methods Mol Biol. 2006;342:129-38. doi: 10.1385/1-59745-123-1:129. Methods Mol Biol. 2006. PMID: 16957372 Review.
-
miRBase: tools for microRNA genomics.Nucleic Acids Res. 2008 Jan;36(Database issue):D154-8. doi: 10.1093/nar/gkm952. Epub 2007 Nov 8. Nucleic Acids Res. 2008. PMID: 17991681 Free PMC article.
-
miROrtho: computational survey of microRNA genes.Nucleic Acids Res. 2009 Jan;37(Database issue):D111-7. doi: 10.1093/nar/gkn707. Epub 2008 Oct 15. Nucleic Acids Res. 2009. PMID: 18927110 Free PMC article.
-
miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D135-9. doi: 10.1093/nar/gkj135. Nucleic Acids Res. 2006. PMID: 16381831 Free PMC article.
-
Computational prediction of microRNA genes.Methods Mol Biol. 2014;1097:437-56. doi: 10.1007/978-1-62703-709-9_20. Methods Mol Biol. 2014. PMID: 24639171 Review.
Cited by
-
Identification of miRNAs and their targets from Brassica napus by high-throughput sequencing and degradome analysis.BMC Genomics. 2012 Aug 24;13:421. doi: 10.1186/1471-2164-13-421. BMC Genomics. 2012. PMID: 22920854 Free PMC article.
-
Functional shifts in insect microRNA evolution.Genome Biol Evol. 2010;2:686-96. doi: 10.1093/gbe/evq053. Epub 2010 Sep 3. Genome Biol Evol. 2010. PMID: 20817720 Free PMC article.
-
MicroRNAs from saliva of anopheline mosquitoes mimic human endogenous miRNAs and may contribute to vector-host-pathogen interactions.Sci Rep. 2019 Feb 27;9(1):2955. doi: 10.1038/s41598-019-39880-1. Sci Rep. 2019. PMID: 30814633 Free PMC article.
-
Evidence for the expression of abundant microRNAs in the locust genome.Sci Rep. 2015 Sep 2;5:13608. doi: 10.1038/srep13608. Sci Rep. 2015. PMID: 26329925 Free PMC article.
-
Large-Scale Annotation and Evolution Analysis of MiRNA in Insects.Genome Biol Evol. 2021 May 7;13(5):evab083. doi: 10.1093/gbe/evab083. Genome Biol Evol. 2021. PMID: 33905491 Free PMC article.
References
-
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foffa R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, Vita GD, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Lauro RD, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

