Replication methods and tools in high-throughput cultivation processes - recognizing potential variations of growth and product formation by on-line monitoring
- PMID: 20233443
- PMCID: PMC2847957
- DOI: 10.1186/1472-6750-10-22
Replication methods and tools in high-throughput cultivation processes - recognizing potential variations of growth and product formation by on-line monitoring
Abstract
Background: High-throughput cultivations in microtiter plates are the method of choice to express proteins from recombinant clone libraries. Such processes typically include several steps, whereby some of them are linked by replication steps: transformation, plating, colony picking, preculture, main culture and induction. In this study, the effects of conventional replication methods and replication tools (8-channel pipette, 96-pin replicators: steel replicator with fixed or spring-loaded pins, plastic replicator with fixed pins) on growth kinetics of Escherichia coli SCS1 pQE-30 pSE111 were observed. Growth was monitored with the BioLector, an on-line monitoring technique for microtiter plates. Furthermore, the influence of these effects on product formation of Escherichia coli pRhotHi-2-EcFbFP was investigated. Finally, a high-throughput cultivation process was simulated with Corynebacterium glutamicum pEKEx2-phoD-GFP, beginning at the colony picking step.
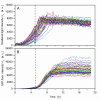
Results: Applying different replication tools and methods for one single strain resulted in high time differences of growth of the slowest and fastest growing culture. The shortest time difference (0.3 h) was evaluated for the 96 cultures that were transferred with an 8-channel pipette from a thawed and mixed cryoculture and the longest time difference (6.9 h) for cultures that were transferred with a steel replicator with fixed pins from a frozen cryoculture. The on-line monitoring of a simulated high-throughput cultivation process revealed strong variances in growth kinetics and a twofold difference in product formation. Another experiment showed that varying growth kinetics, caused by varying initial biomass concentrations (OD(600) of 0.0125 to 0.2) led to strongly varying product formation upon induction at a defined point of time.
Conclusions: To improve the reproducibility of high-throughput cultivation processes and the comparability between different applied cultures, it is strongly recommended to use automated or manual liquid handling stations or, alternatively, multi-channel pipettes. Because of their higher transfer volume and hence precision in comparison to pin replicators, they reduce the variance of initial biomass concentrations. With respect to the results obtained, other methods to increase the comparability between parallel cultivations by compensating differences in biomass concentrations are required, such as using autoinduction media, fed-batch operation of precultures or on-line monitoring in microtiter plates combined with automated liquid handling.
Figures



Similar articles
-
Robo-Lector - a novel platform for automated high-throughput cultivations in microtiter plates with high information content.Microb Cell Fact. 2009 Aug 1;8:42. doi: 10.1186/1475-2859-8-42. Microb Cell Fact. 2009. PMID: 19646274 Free PMC article.
-
Precultures Grown under Fed-Batch Conditions Increase the Reliability and Reproducibility of High-Throughput Screening Results.Biotechnol J. 2019 Nov;14(11):e1800727. doi: 10.1002/biot.201800727. Epub 2019 Aug 5. Biotechnol J. 2019. PMID: 31283111
-
Equalizing growth in high-throughput small scale cultivations via precultures operated in fed-batch mode.Biotechnol Bioeng. 2009 Aug 15;103(6):1095-102. doi: 10.1002/bit.22349. Biotechnol Bioeng. 2009. PMID: 19415772
-
Milliliter-scale stirred tank reactors for the cultivation of microorganisms.Adv Appl Microbiol. 2010;73:61-82. doi: 10.1016/S0065-2164(10)73003-3. Adv Appl Microbiol. 2010. PMID: 20800759 Review.
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
Cited by
-
Screening for optimal protease producing Bacillus licheniformis strains with polymer-based controlled-release fed-batch microtiter plates.Microb Cell Fact. 2021 Feb 23;20(1):51. doi: 10.1186/s12934-021-01541-2. Microb Cell Fact. 2021. PMID: 33622330 Free PMC article.
-
Efficient division and sampling of cell colonies using microcup arrays.Analyst. 2013 Jan 7;138(1):220-8. doi: 10.1039/c2an36065a. Epub 2012 Oct 25. Analyst. 2013. PMID: 23099535 Free PMC article.
-
Optimized Replication of Arrayed Bacterial Mutant Libraries Increases Access to Biological Resources.Microbiol Spectr. 2023 Aug 17;11(4):e0169323. doi: 10.1128/spectrum.01693-23. Epub 2023 Jul 11. Microbiol Spectr. 2023. PMID: 37432110 Free PMC article.
-
Microfabricated arrays for splitting and assay of clonal colonies.Anal Chem. 2012 Dec 18;84(24):10614-20. doi: 10.1021/ac301895t. Epub 2012 Nov 29. Anal Chem. 2012. PMID: 23153031 Free PMC article.
-
Parallel use of shake flask and microtiter plate online measuring devices (RAMOS and BioLector) reduces the number of experiments in laboratory-scale stirred tank bioreactors.J Biol Eng. 2015 May 30;9:9. doi: 10.1186/s13036-015-0005-0. eCollection 2015. J Biol Eng. 2015. PMID: 26265936 Free PMC article.
References
-
- Esposito D, Garvey LA, Chakiath CS. Gateway cloning for protein expression. Methods Mol Biol. 2009;498:31–54. full_text. - PubMed
-
- Duetz WA, Minas W, Kuhner M, Clerval R, Fjallman AHM, Witholt B. Miniaturized microbial growth systems in screening. Bioworld. 2001;2:8–10.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

