Northwestern profiling of potential translation-regulatory proteins in human breast epithelial cells and malignant breast tissues: evidence for pathological activation of the IGF1R IRES
- PMID: 20233590
- PMCID: PMC2868104
- DOI: 10.1016/j.yexmp.2010.03.006
Northwestern profiling of potential translation-regulatory proteins in human breast epithelial cells and malignant breast tissues: evidence for pathological activation of the IGF1R IRES
Abstract
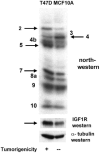
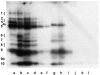
Genes involved in the control of cell proliferation and survival (those genes most important to cancer pathogenesis) are often specifically regulated at the translational level, through RNA-protein interactions involving the 5'-untranslated region of the mRNA. IGF1R is a proto-oncogene strongly implicated in human breast cancer, promoting survival and proliferation of tumor cells, as well as metastasis and chemoresistance. Our lab has focused on the molecular mechanisms regulating IGF1R expression at the translational level. We previously discovered an internal ribosome entry site (IRES) within the 5'-untranslated region of the human IGF1R mRNA, and identified and functionally characterized two individual RNA-binding proteins, HuR and hnRNP C, which bind the IGF1R 5'-UTR and differentially regulate IRES activity. Here we have developed and implemented a high-resolution northwestern profiling strategy to characterize, as a group, the full spectrum of sequence-specific RNA-binding proteins potentially regulating IGF1R translational efficiency through interaction with the 5'-untranslated sequence. The putative IGF1R IRES trans-activating factors (ITAFs) are a heterogeneous group of RNA-binding proteins including hnRNPs originating in the nucleus as well as factors tightly associated with ribosomes in the cytoplasm. The IGF1R ITAFs can be categorized into three distinct groups: (a) high molecular weight external ITAFs, which likely modulate the overall conformation of the 5'-untranslated region of the IGF1R mRNA and thereby the accessibility of the core functional IRES; (b) low molecular weight external ITAFs, which may function as general chaperones to unwind the RNA, and (c) internal ITAFs which may directly facilitate or inhibit the fundamental process of ribosome recruitment to the IRES. We observe dramatic changes in the northwestern profile of non-malignant breast cells downregulating IGF1R expression in association with acinar differentiation in 3-D culture. Most importantly, we are able to assess the RNA-binding activities of these putative translation-regulatory proteins in primary human breast surgical specimens, and begin to discern positive correlations between individual ITAFs and the malignant phenotype. Together with our previous findings, these new data provide further evidence that pathological dysregulation of IGF1R translational control may contribute to development and progression of human breast cancer, and breast metastasis in particular.
Copyright 2010 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures








Similar articles
-
Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells.J Cell Physiol. 2008 Oct;217(1):172-83. doi: 10.1002/jcp.21486. J Cell Physiol. 2008. PMID: 18452152
-
Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein.Mol Cell Biol. 1999 Aug;19(8):5429-40. doi: 10.1128/MCB.19.8.5429. Mol Cell Biol. 1999. PMID: 10409733 Free PMC article.
-
Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs.Mol Cell Biol. 2008 Jan;28(1):40-9. doi: 10.1128/MCB.01298-07. Epub 2007 Oct 29. Mol Cell Biol. 2008. PMID: 17967896 Free PMC article.
-
The role of IRES trans-acting factors in carcinogenesis.Biochim Biophys Acta. 2015 Jul;1849(7):887-97. doi: 10.1016/j.bbagrm.2014.09.012. Epub 2014 Sep 23. Biochim Biophys Acta. 2015. PMID: 25257759 Review.
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
Cited by
-
Small molecule inhibitors of IRES-mediated translation.Cancer Biol Ther. 2015;16(10):1471-85. doi: 10.1080/15384047.2015.1071729. Epub 2015 Jul 15. Cancer Biol Ther. 2015. PMID: 26177060 Free PMC article.
-
Silence of S1 RNA binding domain 1 represses cell growth and promotes apoptosis in human non-small cell lung cancer cells.Transl Lung Cancer Res. 2019 Dec;8(6):760-774. doi: 10.21037/tlcr.2019.10.10. Transl Lung Cancer Res. 2019. PMID: 32010555 Free PMC article.
-
HNRNPC as a candidate biomarker for chemoresistance in gastric cancer.Tumour Biol. 2016 Mar;37(3):3527-34. doi: 10.1007/s13277-015-4144-1. Epub 2015 Oct 9. Tumour Biol. 2016. PMID: 26453116
-
Identification of a five m6A-relevant mRNAs signature and risk score for the prognostication of gastric cancer.J Gastrointest Oncol. 2022 Oct;13(5):2234-2248. doi: 10.21037/jgo-22-962. J Gastrointest Oncol. 2022. PMID: 36388685 Free PMC article.
-
Overexpression of hnRNPC2 induces multinucleation by repression of Aurora B in hepatocellular carcinoma cells.Oncol Lett. 2013 Apr;5(4):1243-1249. doi: 10.3892/ol.2013.1167. Epub 2013 Jan 31. Oncol Lett. 2013. PMID: 23599772 Free PMC article.
References
-
- Belfiore A, Frasca F. IGF and insulin receptor signaling in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13(4):381–406. - PubMed
-
- Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19(38):4437–4440. - PubMed
-
- Chiocchetti A, Gibello L, Carando A, Aspesi A, Secco P, Garelli E, Loreni F, Angelini M, Biava A, Dahl N, Dianzani U, Ramenghi U, Santoro C, Dianzani I. Interactions between RPS19, mutated in Diamond-Blackfan anemia, and the PIM-1 oncoprotein. Haematologica. 2005 Nov;90(11):1453–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

