Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins
- PMID: 20233715
- PMCID: PMC2871492
- DOI: 10.1074/jbc.M109.048785
Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins
Abstract
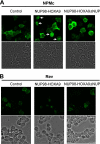
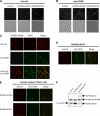
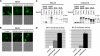
NUP98 is a nucleoporin that plays complex roles in the nucleocytoplasmic trafficking of macromolecules. Rearrangements of the NUP98 gene in human leukemia result in the expression of numerous fusion oncoproteins whose effect on nucleocytoplasmic trafficking is poorly understood. The present study was undertaken to determine the effects of leukemogenic NUP98 fusion proteins on CRM1-mediated nuclear export. NUP98-HOXA9, a prototypic NUP98 fusion, inhibited the nuclear export of two known CRM1 substrates: mutated cytoplasmic nucleophosmin and HIV-1 Rev. In vitro binding assays revealed that NUP98-HOXA9 binds CRM1 through the FG repeat motif in a Ran-GTP-dependent manner similar to but stronger than the interaction between CRM1 and its export substrates. Two NUP98 fusions, NUP98-HOXA9 and NUP98-DDX10, whose fusion partners are structurally and functionally unrelated, interacted with endogenous CRM1 in myeloid cells as shown by co-immunoprecipitation. These leukemogenic NUP98 fusion proteins interacted with CRM1, Ran, and the nucleoporin NUP214 in a manner fundamentally different from that of wild-type NUP98. NUP98-HOXA9 and NUP98-DDX10 formed characteristic aggregates within the nuclei of a myeloid cell line and primary human CD34+ cells and caused aberrant localization of CRM1 to these aggregates. These NUP98 fusions caused nuclear accumulation of two transcription factors, NFAT and NFkappaB, that are regulated by CRM1-mediated export. The nuclear entrapment of NFAT and NFkappaB correlated with enhanced transcription from promoters responsive to these transcription factors. Taken together, the results suggest a new mechanism by which NUP98 fusions dysregulate transcription and cause leukemia, namely, inhibition of CRM1-mediated nuclear export with aberrant nuclear retention of transcriptional regulators.
Figures


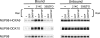
Similar articles
-
The mobile FG nucleoporin Nup98 is a cofactor for Crm1-dependent protein export.Mol Biol Cell. 2010 Jun 1;21(11):1885-96. doi: 10.1091/mbc.e09-12-1041. Epub 2010 Apr 7. Mol Biol Cell. 2010. PMID: 20375145 Free PMC article.
-
The SQSTM1-NUP214 fusion protein interacts with Crm1, activates Hoxa and Meis1 genes, and drives leukemogenesis in mice.PLoS One. 2020 Apr 28;15(4):e0232036. doi: 10.1371/journal.pone.0232036. eCollection 2020. PLoS One. 2020. PMID: 32343715 Free PMC article.
-
Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes.Elife. 2016 Jan 7;5:e09540. doi: 10.7554/eLife.09540. Elife. 2016. PMID: 26740045 Free PMC article.
-
Nucleoporins and nucleocytoplasmic transport in hematologic malignancies.Semin Cancer Biol. 2014 Aug;27:3-10. doi: 10.1016/j.semcancer.2014.02.009. Epub 2014 Mar 18. Semin Cancer Biol. 2014. PMID: 24657637 Review.
-
NUP98 fusion in human leukemia: dysregulation of the nuclear pore and homeodomain proteins.Int J Hematol. 2005 Jul;82(1):21-7. doi: 10.1532/IJH97.04160. Int J Hematol. 2005. PMID: 16105755 Review.
Cited by
-
Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic Transformation.Cancer Discov. 2022 Apr 1;12(4):1152-1169. doi: 10.1158/2159-8290.CD-21-0674. Cancer Discov. 2022. PMID: 34903620 Free PMC article.
-
Regulation of NF-κB Oscillation by Nuclear Transport: Mechanisms Determining the Persistency and Frequency of Oscillation.PLoS One. 2015 Jun 4;10(6):e0127633. doi: 10.1371/journal.pone.0127633. eCollection 2015. PLoS One. 2015. PMID: 26042739 Free PMC article.
-
Nuclear export signal within CALM is necessary for CALM-AF10-induced leukemia.Cancer Sci. 2014 Mar;105(3):315-23. doi: 10.1111/cas.12347. Epub 2014 Feb 13. Cancer Sci. 2014. PMID: 24397609 Free PMC article.
-
Regulation of signal transduction by spatial parameters: a case in NF-κB oscillation.IET Syst Biol. 2015 Apr;9(2):41-51. doi: 10.1049/iet-syb.2013.0020. IET Syst Biol. 2015. PMID: 26672147 Free PMC article.
-
A CALM-derived nuclear export signal is essential for CALM-AF10-mediated leukemogenesis.Blood. 2013 Jun 6;121(23):4758-68. doi: 10.1182/blood-2012-06-435792. Epub 2013 Mar 13. Blood. 2013. PMID: 23487024 Free PMC article.
References
-
- Slape C., Aplan P. D. (2004) Leuk. Lymphoma 45, 1341–1350 - PubMed
-
- Romana S. P., Radford-Weiss I., Ben Abdelali R., Schluth C., Petit A., Dastugue N., Talmant P., Bilhou-Nabera C., Mugneret F., Lafage-Pochitaloff M., Mozziconacci M. J., Andrieu J., Lai J. L., Terre C., Rack K., Cornillet-Lefebvre P., Luquet I., Nadal N., Nguyen-Khac F., Perot C., Van den Akker J., Fert-Ferrer S., Cabrol C., Charrin C., Tigaud I., Poirel H., Vekemans M., Bernard O. A., Berger R. (2006) Leukemia 20, 696–706 - PubMed
-
- Pineault N., Buske C., Feuring-Buske M., Abramovich C., Rosten P., Hogge D. E., Aplan P. D., Humphries R. K. (2003) Blood 101, 4529–4538 - PubMed
-
- Hirose K., Abramovich C., Argiropoulos B., Humphries R. K. (2008) Oncogene 27, 6056–6067 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

