Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair
- PMID: 20233726
- PMCID: PMC2863164
- DOI: 10.1074/jbc.M110.104745
Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair
Abstract
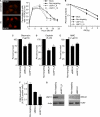
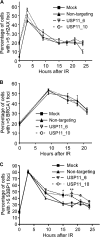
DNA damage repair and checkpoint responses prevent genome instability and provide a barrier to the development of cancer. Inherited mutations in DNA damage response (DDR) genes such as those that encode the homologous recombination (HR) proteins BRCA1 and BRCA2 cause cancer predisposition syndromes. PARP inhibitors are an exciting new class of targeted therapy for treating patients with HR repair-defective tumors. In this study, we use an RNAi screen to identify genes that when silenced cause synthetic lethality with the PARP inhibitor AZD2281. This screen identified the deubiquitylating enzyme USP11 as a participant in HR repair of DNA double-strand breaks. Silencing USP11 with siRNA leads to spontaneous DDR activation in otherwise undamaged cells and hypersensitivity to PARP inhibition, ionizing radiation, and other genotoxic stress agents. Moreover, we demonstrate that HR repair is defective in USP11-silenced cells. Finally, the recruitment of a subset of double-strand break repair proteins including RAD51 and 53BP1 to repair foci is misregulated in the absence of USP11 catalytic activity. Thus, our synthetic lethal approach identified USP11 as a component of the HR double-strand break repair pathway.
Figures

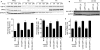
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

