Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma
- PMID: 20233883
- PMCID: PMC2853253
- DOI: 10.1158/1078-0432.CCR-09-2879
Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma
Abstract
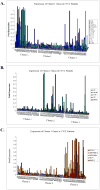
Purpose: Average survival of cutaneous T-cell lymphoma (CTCL) is associated with clinical stage at diagnosis, where stage I has a favorable survival prognosis, whereas patients with more advanced stages succumb to their disease within 5 years. Although the majority of patients present with an early-stage CTCL, 15% to 20% of them will inevitably progress. Current state-of-the-art clinical criteria cannot identify individuals with stage I disease who are at risk of progression. The purpose of the current work is to gain novel molecular insight into the pathophysiology of CTCL to be able to identify patients with poor versus favorable prognosis. Our previous work used microarray analysis of skin biopsies from 62 CTCL patients to perform an unsupervised analysis of gene expression, which revealed three distinct transcription profile clusters.
Experimental design: In the present study, we used reverse transcription-PCR to confirm gene expression levels for a subset of representative genes in each cluster. We also performed a Kaplan-Meier analysis of survival and disease progression based on the 6 years of clinical follow-up.
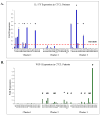
Results: Our reverse transcription-PCR results confirmed the upregulation of representative genes for each cluster, whereas clinical analysis documents that all stage I cases that progressed to stage II and beyond were in poor and intermediate prognosis clusters 1 and 3 and none were in favorable prognosis cluster 2. This analysis also identified certain genes that were preferentially expressed in favorable (e.g., WIF-1) versus poor (e.g., IL-17F) prognosis clusters.
Conclusion: This work suggests that it may be possible to stratify CTCL patients into low-risk, intermediate-risk, and high-risk groups based on gene expression.
Copyright 2010 AACR.
Conflict of interest statement
Figures

Similar articles
-
Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome.Blood. 2007 Oct 15;110(8):3015-27. doi: 10.1182/blood-2006-12-061507. Epub 2007 Jul 16. Blood. 2007. PMID: 17638852 Free PMC article.
-
The Use of Transcriptional Profiling to Improve Personalized Diagnosis and Management of Cutaneous T-cell Lymphoma (CTCL).Clin Cancer Res. 2015 Jun 15;21(12):2820-9. doi: 10.1158/1078-0432.CCR-14-3322. Epub 2015 Mar 16. Clin Cancer Res. 2015. PMID: 25779945 Free PMC article.
-
Single-Cell Profiling of Cutaneous T-Cell Lymphoma Reveals Underlying Heterogeneity Associated with Disease Progression.Clin Cancer Res. 2019 May 15;25(10):2996-3005. doi: 10.1158/1078-0432.CCR-18-3309. Epub 2019 Feb 4. Clin Cancer Res. 2019. PMID: 30718356 Free PMC article.
-
Current status of cutaneous T-cell lymphoma: molecular diagnosis, pathogenesis, therapy and future directions.Onkologie. 2003 Oct;26(5):477-83. doi: 10.1159/000072099. Onkologie. 2003. PMID: 14605466 Review.
-
Genetics of Cutaneous T Cell Lymphoma: From Bench to Bedside.Curr Treat Options Oncol. 2016 Jul;17(7):33. doi: 10.1007/s11864-016-0410-8. Curr Treat Options Oncol. 2016. PMID: 27262707 Review.
Cited by
-
The changing therapeutic landscape, burden of disease, and unmet needs in patients with cutaneous T-cell lymphoma.Br J Haematol. 2021 Feb;192(4):683-696. doi: 10.1111/bjh.17117. Epub 2020 Oct 23. Br J Haematol. 2021. PMID: 33095448 Free PMC article. Review.
-
Understanding Cell Lines, Patient-Derived Xenograft and Genetically Engineered Mouse Models Used to Study Cutaneous T-Cell Lymphoma.Cells. 2022 Feb 9;11(4):593. doi: 10.3390/cells11040593. Cells. 2022. PMID: 35203244 Free PMC article. Review.
-
Malignant T cells express lymphotoxin α and drive endothelial activation in cutaneous T cell lymphoma.Oncotarget. 2015 Jun 20;6(17):15235-49. doi: 10.18632/oncotarget.3837. Oncotarget. 2015. PMID: 25915535 Free PMC article.
-
Genetic markers associated with progression in early mycosis fungoides.J Eur Acad Dermatol Venereol. 2014 Nov;28(11):1431-5. doi: 10.1111/jdv.12299. Epub 2013 Oct 31. J Eur Acad Dermatol Venereol. 2014. PMID: 24171863 Free PMC article.
-
Identification of geographic clustering and regions spared by cutaneous T-cell lymphoma in Texas using 2 distinct cancer registries.Cancer. 2015 Jun 15;121(12):1993-2003. doi: 10.1002/cncr.29301. Epub 2015 Feb 27. Cancer. 2015. PMID: 25728286 Free PMC article.
References
-
- Lamberg SI, Bunn PA., Jr Cutaneous T-cell lymphomas. Summary of the Mycosis Fungoides Cooperative Group-National Cancer Institute Workshop. Arch Dermatol. 1979;115:1103–5. - PubMed
-
- Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973–2002. Arch Dermatol. 2007;143:854–9. - PubMed
-
- Siegel RS, Pandolfino T, Guitart J, Rosen S, Kuzel TM. Primary cutaneous T-cell lymphoma: review and current concepts. J Clin Oncol. 2000;18:2908–25. - PubMed
-
- Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105:3768–85. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

