The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair
- PMID: 20233923
- PMCID: PMC2876487
- DOI: 10.1128/JB.01266-09
The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair
Abstract
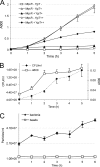
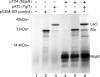
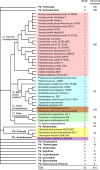
Toxin-antitoxin (TA) systems are plasmid- or chromosome-encoded protein complexes composed of a stable toxin and a short-lived inhibitor of the toxin. In cultures of Escherichia coli, transcription of toxin-antitoxin genes was induced in a nondividing subpopulation of bacteria that was tolerant to bactericidal antibiotics. Along with transcription of known toxin-antitoxin operons, transcription of mqsR and ygiT, two adjacent genes with multiple TA-like features, was induced in this cell population. Here we show that mqsR and ygiT encode a toxin-antitoxin system belonging to a completely new family which is represented in several groups of bacteria. The mqsR gene encodes a toxin, and ectopic expression of this gene inhibits growth and induces rapid shutdown of protein synthesis in vivo. ygiT encodes an antitoxin, which protects cells from the effects of MqsR. These two genes constitute a single operon which is transcriptionally repressed by the product of ygiT. We confirmed that transcription of this operon is induced in the ampicillin-tolerant fraction of a growing population of E. coli and in response to activation of the HipA toxin. Expression of the MqsR toxin does not kill bacteria but causes reversible growth inhibition and elongation of cells.
Figures



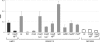
Similar articles
-
Activation of Toxin-Antitoxin System Toxins Suppresses Lethality Caused by the Loss of σE in Escherichia coli.J Bacteriol. 2015 Jul;197(14):2316-24. doi: 10.1128/JB.00079-15. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917909 Free PMC article.
-
Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module.FEMS Microbiol Lett. 2007 Mar;268(1):112-9. doi: 10.1111/j.1574-6968.2006.00563.x. FEMS Microbiol Lett. 2007. PMID: 17263853
-
Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD.Environ Microbiol. 2010 May;12(5):1105-21. doi: 10.1111/j.1462-2920.2009.02147.x. Epub 2010 Jan 26. Environ Microbiol. 2010. PMID: 20105222 Free PMC article.
-
Regulation of toxin-antitoxin systems by proteolysis.Plasmid. 2013 Jul;70(1):33-41. doi: 10.1016/j.plasmid.2013.01.007. Epub 2013 Feb 8. Plasmid. 2013. PMID: 23396045 Review.
-
Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response.Appl Environ Microbiol. 2011 Aug 15;77(16):5577-83. doi: 10.1128/AEM.05068-11. Epub 2011 Jun 17. Appl Environ Microbiol. 2011. PMID: 21685157 Free PMC article. Review.
Cited by
-
Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis.Microb Biotechnol. 2015 Nov;8(6):961-73. doi: 10.1111/1751-7915.12294. Epub 2015 Jun 25. Microb Biotechnol. 2015. PMID: 26112399 Free PMC article.
-
Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation.J Biol Chem. 2011 Jan 21;286(3):2285-96. doi: 10.1074/jbc.M110.172643. Epub 2010 Nov 9. J Biol Chem. 2011. PMID: 21068382 Free PMC article.
-
Crystallization of the HigBA2 toxin-antitoxin complex from Vibrio cholerae.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Sep;69(Pt 9):1052-9. doi: 10.1107/S1744309113021490. Epub 2013 Aug 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23989162 Free PMC article.
-
Effect of ZnO nanoparticles on biofilm formation and gene expression of the toxin-antitoxin system in clinical isolates of Pseudomonas aeruginosa.Ann Clin Microbiol Antimicrob. 2023 Oct 5;22(1):89. doi: 10.1186/s12941-023-00639-2. Ann Clin Microbiol Antimicrob. 2023. PMID: 37798613 Free PMC article.
-
Integrative biology of persister cell formation: molecular circuitry, phenotypic diversification and fitness effects.J R Soc Interface. 2022 Sep;19(194):20220129. doi: 10.1098/rsif.2022.0129. Epub 2022 Sep 14. J R Soc Interface. 2022. PMID: 36099930 Free PMC article. Review.
References
-
- Allen, G. C., Jr., and A. Kornberg. 1991. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem. 266:22096-22101. - PubMed
-
- Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

