Regulation of aerobic and anaerobic D-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (YeaT)
- PMID: 20233924
- PMCID: PMC2863561
- DOI: 10.1128/JB.01665-09
Regulation of aerobic and anaerobic D-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (YeaT)
Abstract
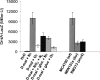
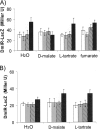
Escherichia coli K-12 is able to grow under aerobic conditions on D-malate using DctA for D-malate uptake and the D-malate dehydrogenase DmlA (formerly YeaU) for converting D-malate to pyruvate. Induction of dmlA encoding DmlA required an intact dmlR (formerly yeaT) gene, which encodes DmlR, a LysR-type transcriptional regulator. Induction of dmlA by DmlR required the presence of D-malate or L- or meso-tartrate, but only D-malate supported aerobic growth. The regulator of general C(4)-dicarboxylate metabolism (DcuS-DcuR two-component system) had some effect on dmlA expression. The anaerobic L-tartrate regulator TtdR or the oxygen sensors ArcB-ArcA and FNR did not have a major effect on dmlA expression. DmlR has a high level of sequence identity (49%) with TtdR, the L- and meso-tartrate-specific regulator of L-tartrate fermentation in E. coli. dmlA was also expressed at high levels under anaerobic conditions, and the bacteria had D-malate dehydrogenase activity. These bacteria, however, were not able to grow on D-malate since the anaerobic pathway for D-malate degradation has a predicted yield of < or = 0 ATP/mol D-malate. Slow anaerobic growth on D-malate was observed when glycerol was also provided as an electron donor, and D-malate was used in fumarate respiration. The expression of dmlR is subject to negative autoregulation. The network for regulation and coordination of the central and peripheral pathways for C(4)-dicarboxylate metabolism by the regulators DcuS-DcuR, DmlR, and TtdR is discussed.
Figures


Similar articles
-
Regulation of tartrate metabolism by TtdR and relation to the DcuS-DcuR-regulated C4-dicarboxylate metabolism of Escherichia coli.Microbiology (Reading). 2009 Nov;155(Pt 11):3632-3640. doi: 10.1099/mic.0.031401-0. Epub 2009 Aug 6. Microbiology (Reading). 2009. PMID: 19661178
-
C4-Dicarboxylate Utilization in Aerobic and Anaerobic Growth.EcoSal Plus. 2016 Jun;7(1):10.1128/ecosalplus.ESP-0021-2015. doi: 10.1128/ecosalplus.ESP-0021-2015. EcoSal Plus. 2016. PMID: 27415771 Free PMC article.
-
Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli.J Bacteriol. 1999 Feb;181(4):1238-48. doi: 10.1128/JB.181.4.1238-1248.1999. J Bacteriol. 1999. PMID: 9973351 Free PMC article.
-
Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors.Biochim Biophys Acta. 1997 Jul 4;1320(3):217-34. doi: 10.1016/s0005-2728(97)00034-0. Biochim Biophys Acta. 1997. PMID: 9230919 Review.
-
Sensing by the membrane-bound sensor kinase DcuS: exogenous versus endogenous sensing of C(4)-dicarboxylates in bacteria.Future Microbiol. 2010 Sep;5(9):1383-402. doi: 10.2217/fmb.10.103. Future Microbiol. 2010. PMID: 20860483 Review.
Cited by
-
The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists.Sci Rep. 2018 Feb 1;8(1):2102. doi: 10.1038/s41598-018-20283-7. Sci Rep. 2018. PMID: 29391435 Free PMC article.
-
Multiple turnovers of the nicotino-enzyme PdxB require α-keto acids as cosubstrates.Biochemistry. 2010 Nov 2;49(43):9249-55. doi: 10.1021/bi101291d. Biochemistry. 2010. PMID: 20831184 Free PMC article.
-
Phosphoproteome Study of Escherichia coli Devoid of Ser/Thr Kinase YeaG During the Metabolic Shift From Glucose to Malate.Front Microbiol. 2021 Apr 6;12:657562. doi: 10.3389/fmicb.2021.657562. eCollection 2021. Front Microbiol. 2021. PMID: 33889145 Free PMC article.
-
d-2-Hydroxyglutarate dehydrogenase plays a dual role in l-serine biosynthesis and d-malate utilization in the bacterium Pseudomonas stutzeri.J Biol Chem. 2018 Oct 5;293(40):15513-15523. doi: 10.1074/jbc.RA118.003897. Epub 2018 Aug 21. J Biol Chem. 2018. PMID: 30131334 Free PMC article.
-
Acid Evolution of Escherichia coli K-12 Eliminates Amino Acid Decarboxylases and Reregulates Catabolism.Appl Environ Microbiol. 2017 May 31;83(12):e00442-17. doi: 10.1128/AEM.00442-17. Print 2017 Jun 15. Appl Environ Microbiol. 2017. PMID: 28389540 Free PMC article.
References
-
- Blumer, C., A. Kleefeld, D. Lehnen, M. Heintz, U. Dobrindt, G. Nagy, K. Michaelis, L. Emödy, T. Polen, R. Rachel, V. F. Wendisch, and G. Unden. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287-3298. - PubMed
-
- Bongaerts, J., S. Zoske, U. Weidner, and G. Unden. 1995. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol. Microbiol. 16:521-534. - PubMed
-
- Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

