Repellent taxis in response to nickel ion requires neither Ni2+ transport nor the periplasmic NikA binding protein
- PMID: 20233931
- PMCID: PMC2863559
- DOI: 10.1128/JB.00854-09
Repellent taxis in response to nickel ion requires neither Ni2+ transport nor the periplasmic NikA binding protein
Erratum in
- J Bacteriol. 2010 Aug;192(16):4259
Abstract
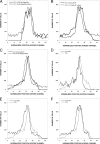
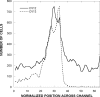
Ni(2+) and Co(2+) are sensed as repellents by the Escherichia coli Tar chemoreceptor. The periplasmic Ni(2+) binding protein, NikA, has been suggested to sense Ni(2+). We show here that neither NikA nor the membrane-bound NikB and NikC proteins of the Ni(2+) transport system are required for repellent taxis in response to Ni(2+).
Figures

Similar articles
-
Histidine 416 of the periplasmic binding protein NikA is essential for nickel uptake in Escherichia coli.FEBS Lett. 2011 Feb 18;585(4):711-5. doi: 10.1016/j.febslet.2011.01.038. Epub 2011 Feb 1. FEBS Lett. 2011. PMID: 21281641
-
Purification and characterization of the periplasmic nickel-binding protein NikA of Escherichia coli K12.Eur J Biochem. 1995 Feb 1;227(3):857-65. doi: 10.1111/j.1432-1033.1995.tb20211.x. Eur J Biochem. 1995. PMID: 7867647
-
Biochemical studies highlight determinants for metal selectivity in the Escherichia coli periplasmic solute binding protein NikA.Metallomics. 2022 Nov 17;14(11):mfac084. doi: 10.1093/mtomcs/mfac084. Metallomics. 2022. PMID: 36255398 Free PMC article.
-
Nickel transport systems in microorganisms.Arch Microbiol. 2000 Jan;173(1):1-9. doi: 10.1007/s002030050001. Arch Microbiol. 2000. PMID: 10648098 Review.
-
Sinorhizobial chemotaxis: a departure from the enterobacterial paradigm.Microbiology (Reading). 2002 Mar;148(Pt 3):627-631. doi: 10.1099/00221287-148-3-627. Microbiology (Reading). 2002. PMID: 11882696 Review. No abstract available.
Cited by
-
Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins.Proc Natl Acad Sci U S A. 2012 Mar 6;109(10):3766-71. doi: 10.1073/pnas.1115719109. Epub 2012 Feb 21. Proc Natl Acad Sci U S A. 2012. PMID: 22355139 Free PMC article.
-
Dose-Response Analysis of Chemotactic Signaling Response in Salmonella typhimurium LT2 upon Exposure to Cysteine/Cystine Redox Pair.PLoS One. 2016 Apr 7;11(4):e0152815. doi: 10.1371/journal.pone.0152815. eCollection 2016. PLoS One. 2016. PMID: 27054963 Free PMC article.
-
Enhancement of Swimming Speed Leads to a More-Efficient Chemotactic Response to Repellent.Appl Environ Microbiol. 2015 Dec 11;82(4):1205-1214. doi: 10.1128/AEM.03397-15. Print 2016 Feb 15. Appl Environ Microbiol. 2015. PMID: 26655753 Free PMC article.
-
Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling.Elife. 2014 Mar 25;3:e02151. doi: 10.7554/eLife.02151. Elife. 2014. PMID: 24668172 Free PMC article.
-
Multiple detection of both attractants and repellents by the dCache-chemoreceptor SO_1056 of Shewanella oneidensis.FEBS J. 2022 Nov;289(21):6752-6766. doi: 10.1111/febs.16548. Epub 2022 Jun 24. FEBS J. 2022. PMID: 35668695 Free PMC article.
References
-
- Abouhamad, W. N., M. Manson, M. M. Gibson, and C. F. Higgins. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035-1047. - PubMed
-
- Abouhamad, W. N., and M. D. Manson. 1994. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 14:1077-1092. - PubMed
-
- Aksamit, R. R., and D. E. Koshland, Jr. 1974. Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry 13:4473-4478. - PubMed
-
- Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

