Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa
- PMID: 20233934
- PMCID: PMC2863569
- DOI: 10.1128/JB.01528-09
Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa
Abstract
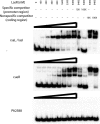
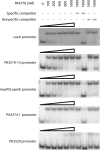
The LasR/LasI quorum-sensing system in Pseudomonas aeruginosa influences global gene expression and mediates pathogenesis. In this study, we show that the quorum-sensing system activates, via the transcriptional regulator PA4778, a copper resistance system composed of 11 genes. The quorum-sensing global regulator LasR was recently shown to directly activate transcription of PA4778, a cueR homolog and a MerR-type transcriptional regulator. Using molecular genetic methods and bioinformatics, we verify the interaction of LasR with the PA4778 promoter and further demonstrate the LasR binding site. We also identify a putative PA4778 binding motif and show that the protein directly binds to and activates five promoters controlling the expression of 11 genes--PA3519 to -15, PA3520, mexPQ-opmE, PA3574.1, and cueA, a virulence factor in a murine model. Using gene disruptions, we show that PA4778, along with 7 of 11 gene targets of PA4778, increases the sensitivity of P. aeruginosa to elevated copper concentrations. This work identifies a cellular function for PA4778 and four other previously unannotated genes (PA3515, PA3516, PA3517, and PA3518) and suggests a potential role for copper in the quorum response. We propose to name PA4778 cueR.
Figures
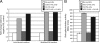
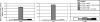




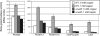
References
-
- Abramoff, M. D., P. J. Magalhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-41.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. - PubMed
-
- Ansari, A. Z., M. L. Chael, and T. V. O'Halloran. 1992. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 355:87-89. - PubMed
-
- Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

