PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock
- PMID: 20233950
- PMCID: PMC2861452
- DOI: 10.1105/tpc.109.072892
PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock
Abstract
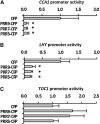
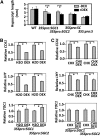
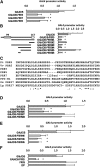
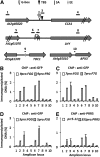
An interlocking transcriptional-translational feedback loop of clock-associated genes is thought to be the central oscillator of the circadian clock in plants. TIMING OF CAB EXPRESSION1 (also called PSEUDO-RESPONSE REGULATOR1 [PRR1]) and two MYB transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), play pivotal roles in the loop. Genetic studies have suggested that PRR9, PRR7, and PRR5 also act within or close to the loop; however, their molecular functions remain unknown. Here, we demonstrate that PRR9, PRR7, and PRR5 act as transcriptional repressors of CCA1 and LHY. PRR9, PRR7, and PRR5 each suppress CCA1 and LHY promoter activities and confer transcriptional repressor activity to a heterologous DNA binding protein in a transient reporter assay. Using a glucocorticoid-induced PRR5-GR (glucorticoid receptor) construct, we found that PRR5 directly downregulates CCA1 and LHY expression. Furthermore, PRR9, PRR7, and PRR5 associate with the CCA1 and LHY promoters in vivo, coincident with the timing of decreased CCA1 and LHY expression. These results suggest that the repressor activities of PRR9, PRR7, and PRR5 on the CCA1 and LHY promoter regions constitute the molecular mechanism that accounts for the role of these proteins in the feedback loop of the circadian clock.
Figures


References
-
- Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316: 1194–1199
-
- Eriksson M.E., Hanano S., Southern M.M., Hall A., Millar A.J. (2003). Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218: 159–162 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

