An optimized TOPS+ comparison method for enhanced TOPS models
- PMID: 20236520
- PMCID: PMC2858036
- DOI: 10.1186/1471-2105-11-138
An optimized TOPS+ comparison method for enhanced TOPS models
Abstract
Background: Although methods based on highly abstract descriptions of protein structures, such as VAST and TOPS, can perform very fast protein structure comparison, the results can lack a high degree of biological significance. Previously we have discussed the basic mechanisms of our novel method for structure comparison based on our TOPS+ model (Topological descriptions of Protein Structures Enhanced with Ligand Information). In this paper we show how these results can be significantly improved using parameter optimization, and we call the resulting optimised TOPS+ method as advanced TOPS+ comparison method i.e. advTOPS+.
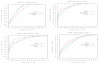
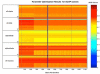
Results: We have developed a TOPS+ string model as an improvement to the TOPS 123 graph model by considering loops as secondary structure elements (SSEs) in addition to helices and strands, representing ligands as first class objects, and describing interactions between SSEs, and SSEs and ligands, by incoming and outgoing arcs, annotating SSEs with the interaction direction and type. Benchmarking results of an all-against-all pairwise comparison using a large dataset of 2,620 non-redundant structures from the PDB40 dataset 4 demonstrate the biological significance, in terms of SCOP classification at the superfamily level, of our TOPS+ comparison method.
Conclusions: Our advanced TOPS+ comparison shows better performance on the PDB40 dataset 4 compared to our basic TOPS+ method, giving 90% accuracy for SCOP alpha+beta; a 6% increase in accuracy compared to the TOPS and basic TOPS+ methods. It also outperforms the TOPS, basic TOPS+ and SSAP comparison methods on the Chew-Kedem dataset 5, achieving 98% accuracy.
Software availability: The TOPS+ comparison server is available at http://balabio.dcs.gla.ac.uk/mallika/WebTOPS/.
Figures


Similar articles
-
A novel method for comparing topological models of protein structures enhanced with ligand information.Bioinformatics. 2008 Dec 1;24(23):2698-705. doi: 10.1093/bioinformatics/btn518. Epub 2008 Oct 7. Bioinformatics. 2008. PMID: 18842602
-
TOPS: an enhanced database of protein structural topology.Nucleic Acids Res. 2004 Jan 1;32(Database issue):D251-4. doi: 10.1093/nar/gkh060. Nucleic Acids Res. 2004. PMID: 14681405 Free PMC article.
-
Protein structure topological comparison, discovery and matching service.Bioinformatics. 2005 May 15;21(10):2537-8. doi: 10.1093/bioinformatics/bti331. Epub 2005 Mar 1. Bioinformatics. 2005. PMID: 15741246
-
A new method to analyze the similarity of protein structure using TOPS representations.J Biomol Struct Dyn. 2008 Dec;26(3):367-74. doi: 10.1080/07391102.2008.10507251. J Biomol Struct Dyn. 2008. PMID: 18808202
-
Identification of local variations within secondary structures of proteins.Acta Crystallogr D Biol Crystallogr. 2015 May;71(Pt 5):1077-86. doi: 10.1107/S1399004715003144. Epub 2015 Apr 24. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25945573
Cited by
-
Efficient Multicriteria Protein Structure Comparison on Modern Processor Architectures.Biomed Res Int. 2015;2015:563674. doi: 10.1155/2015/563674. Epub 2015 Oct 28. Biomed Res Int. 2015. PMID: 26605332 Free PMC article.
References
-
- Gilbert D, Westhead DR, Viksna J. In: Artificial Intelligence and Heuristic Methods in Bioinformatics, Volume 183 of NATO Science Series: Computer & Systems Sciences. Frasconi P, Shamir R, editor. IOS Press; 2003. Techniques for Comparison, Pattern Matching and Pattern Discovery: From Sequences to Protein Topology; pp. 128–147.
-
- Viksna J, Gilbert D. Algorithms in BioInformatics, Volume 2149 of Lecture Notes in Comput. Sci. Springer-Verlag; 2001. Pattern Matching and Pattern Discovery Algorithms for Protein Topologies; pp. 98–111.
-
- Chew LP, Kedem K. Finding the Consensus Shape for a Protein Family. Algorithmica. 2003;38:115–129. doi: 10.1007/s00453-003-1045-2. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

