Solution structure of a Plasmodium falciparum AMA-1/MSP 1 chimeric protein vaccine candidate (PfCP-2.9) for malaria
- PMID: 20236549
- PMCID: PMC2850360
- DOI: 10.1186/1475-2875-9-76
Solution structure of a Plasmodium falciparum AMA-1/MSP 1 chimeric protein vaccine candidate (PfCP-2.9) for malaria
Abstract
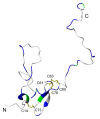
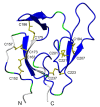
Background: The Plasmodium falciparum chimeric protein PfCP-2.9 is a promising asexual-stage malaria vaccine evaluated in clinical trials. This chimeric protein consists of two cysteine-rich domains: domain III of the apical membrane antigen 1 (AMA-1 [III]) and the C-terminal region of the merozoite surface protein 1 (MSP1-19). It has been reported that the fusion of these two antigens enhanced their immunogenicity and antibody-mediated inhibition of parasite growth in vitro.
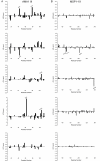
Methods: The 15N-labeled and 13C/15N-labeled PfCP-2.9 was produced in Pichia pastoris for nuclear magnetic resonance (NMR) structure analysis. The chemical shift assignments of PfCP-2.9 were compared with those previously reported for the individual domains (i.e., PfAMA-1(III) or PfMSP 1-19). The two-dimensional spectra and transverse relaxation rates (R2) of the PfMSP1-19 alone were compared with that of the PfCP-2.9.
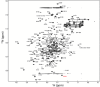
Results: Confident backbone assignments were obtained for 122 out of 241 residues of PfCP-2.9. The assigned residues in PfCP-2.9 were very similar to those previously reported for the individual domains. The conformation of the PfMSP1-19 in different constructs is essentially the same. Comparison of transverse relaxation rates (R2) strongly suggests no weak interaction between the domains.
Conclusions: These data indicate that the fusion of AMA-1(III) and MSP1-19 as chimeric protein did not change their structures, supporting the use of the chimeric protein as a potential malaria vaccine.
Figures


Similar articles
-
Epitope mapping of PfCP-2.9, an asexual blood-stage vaccine candidate of Plasmodium falciparum.Malar J. 2010 Apr 12;9:94. doi: 10.1186/1475-2875-9-94. Malar J. 2010. PMID: 20384992 Free PMC article.
-
Stability and potency of the Plasmodium falciparum MSP1-19/AMA-1(III) chimeric vaccine candidate with Montanide ISA720 adjuvant.Vaccine. 2010 Apr 19;28(18):3152-8. doi: 10.1016/j.vaccine.2010.02.054. Epub 2010 Mar 1. Vaccine. 2010. PMID: 20197139
-
Safety and immunogenicity of a malaria vaccine, Plasmodium falciparum AMA-1/MSP-1 chimeric protein formulated in montanide ISA 720 in healthy adults.PLoS One. 2008 Apr 9;3(4):e1952. doi: 10.1371/journal.pone.0001952. PLoS One. 2008. PMID: 18398475 Free PMC article. Clinical Trial.
-
Fusion of two malaria vaccine candidate antigens enhances product yield, immunogenicity, and antibody-mediated inhibition of parasite growth in vitro.J Immunol. 2004 May 15;172(10):6167-74. doi: 10.4049/jimmunol.172.10.6167. J Immunol. 2004. PMID: 15128804
-
Glycobiology of Plasmodium falciparum: an emerging area of research.Glycoconj J. 1996 Feb;13(1):1-3. doi: 10.1007/BF01049673. Glycoconj J. 1996. PMID: 8785480 Review. No abstract available.
Cited by
-
O-GlcNAc modification of the anti-malarial vaccine candidate PfAMA1: in silico-defined structural changes and potential to generate a better vaccine.Mol Biol Rep. 2012 Apr;39(4):4663-72. doi: 10.1007/s11033-011-1258-4. Epub 2011 Oct 22. Mol Biol Rep. 2012. PMID: 22020851
-
Vaccines against a Major Cause of Abortion in Cattle, Neospora caninum Infection.Animals (Basel). 2011 Sep 8;1(3):306-25. doi: 10.3390/ani1030306. Animals (Basel). 2011. PMID: 26486502 Free PMC article. Review.
-
Efficient production of (2)H, (13)C, (15)N-enriched industrial enzyme Rhizopus chinensis lipase with native disulfide bonds.Microb Cell Fact. 2016 Jul 13;15(1):123. doi: 10.1186/s12934-016-0522-7. Microb Cell Fact. 2016. PMID: 27411547 Free PMC article.
-
Epitope mapping of PfCP-2.9, an asexual blood-stage vaccine candidate of Plasmodium falciparum.Malar J. 2010 Apr 12;9:94. doi: 10.1186/1475-2875-9-94. Malar J. 2010. PMID: 20384992 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

