Binding of the ClpA unfoldase opens the axial gate of ClpP peptidase
- PMID: 20236930
- PMCID: PMC2863180
- DOI: 10.1074/jbc.M109.090498
Binding of the ClpA unfoldase opens the axial gate of ClpP peptidase
Abstract
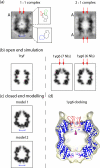
ClpP is a serine protease whose active sites are sequestered in a cavity enclosed between two heptameric rings of subunits. The ability of ClpP to process folded protein substrates depends on its being partnered by an AAA+ ATPase/unfoldase, ClpA or ClpX. In active complexes, substrates are unfolded and fed along an axial channel to the degradation chamber inside ClpP. We have used cryoelectron microscopy at approximately 11-A resolution to investigate the three-dimensional structure of ClpP complexed with either one or two end-mounted ClpA hexamers. In the absence of ClpA, the apical region of ClpP is sealed; however, it opens on ClpA binding, creating an access channel. This region is occupied by the N-terminal loops (residues 1-17) of ClpP, which tend to be poorly visible in crystal structures, indicative of conformational variability. Nevertheless, we were able to model the closed-to-open transition that accompanies ClpA binding in terms of movements of these loops; in particular, "up" conformations of the loops correlate with the open state. The main part of ClpP, the barrel formed by 14 copies of residues 18-193, is essentially unchanged by the interaction with ClpA. Using difference mapping, we localized the binding site for ClpA to a peripheral pocket between adjacent ClpP subunits. Based on these observations, we propose that access to the ClpP degradation chamber is controlled allosterically by hinged movements of its N-terminal loops, which the symmetry-mismatched binding of ClpA suffices to induce.
Figures



Similar articles
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
-
E. coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP.J Mol Biol. 2013 Aug 9;425(15):2795-812. doi: 10.1016/j.jmb.2013.04.019. Epub 2013 Apr 29. J Mol Biol. 2013. PMID: 23639359 Free PMC article.
-
The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation.J Biol Chem. 2005 Apr 22;280(16):16185-96. doi: 10.1074/jbc.M414124200. Epub 2005 Feb 8. J Biol Chem. 2005. PMID: 15701650
-
The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes.J Struct Biol. 2006 Feb;153(2):113-28. doi: 10.1016/j.jsb.2005.09.011. Epub 2005 Dec 1. J Struct Biol. 2006. PMID: 16406682 Free PMC article.
-
Dynamics of the ClpP serine protease: a model for self-compartmentalized proteases.Crit Rev Biochem Mol Biol. 2014 Sep-Oct;49(5):400-12. doi: 10.3109/10409238.2014.925421. Epub 2014 Jun 10. Crit Rev Biochem Mol Biol. 2014. PMID: 24915503 Review.
Cited by
-
Archaeal proteasomes and sampylation.Subcell Biochem. 2013;66:297-327. doi: 10.1007/978-94-007-5940-4_11. Subcell Biochem. 2013. PMID: 23479445 Free PMC article. Review.
-
Reversible inhibition of the ClpP protease via an N-terminal conformational switch.Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):E6447-E6456. doi: 10.1073/pnas.1805125115. Epub 2018 Jun 25. Proc Natl Acad Sci U S A. 2018. PMID: 29941580 Free PMC article.
-
Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.J Biol Chem. 2016 Apr 1;291(14):7465-76. doi: 10.1074/jbc.M115.700344. Epub 2016 Feb 8. J Biol Chem. 2016. PMID: 26858247 Free PMC article.
-
Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics.J Biol Chem. 2011 Oct 28;286(43):37590-601. doi: 10.1074/jbc.M111.277848. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21900233 Free PMC article.
-
Disordered proteinaceous machines.Chem Rev. 2014 Jul 9;114(13):6806-43. doi: 10.1021/cr4007329. Epub 2014 Apr 4. Chem Rev. 2014. PMID: 24702702 Free PMC article. Review. No abstract available.
References
-
- Wickner S., Maurizi M. R., Gottesman S. (1999) Science 286, 1888–1893 - PubMed
-
- Striebel F., Kress W., Weber-Ban E. (2009) Curr. Opin. Struct. Biol. 19, 209–217 - PubMed
-
- Gottesman S. (2003) Annu. Rev. Cell Dev. Biol. 19, 565–587 - PubMed
-
- Snider J., Houry W. A. (2008) Biochem. Soc. Trans. 36, 72–77 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

