Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology
- PMID: 20236988
- PMCID: PMC2860132
- DOI: 10.1093/nar/gkq164
Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology
Abstract
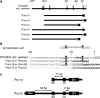
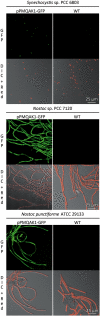
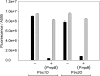
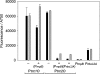
Cyanobacteria are suitable for sustainable, solar-powered biotechnological applications. Synthetic biology connects biology with computational design and an engineering perspective, but requires efficient tools and information about the function of biological parts and systems. To enable the development of cyanobacterial Synthetic Biology, several molecular tools were developed and characterized: (i) a broad-host-range BioBrick shuttle vector, pPMQAK1, was constructed and confirmed to replicate in Escherichia coli and three different cyanobacterial strains. (ii) The fluorescent proteins Cerulean, GFPmut3B and EYFP have been demonstrated to work as reporter proteins in cyanobacteria, in spite of the strong background of photosynthetic pigments. (iii) Several promoters, like P(rnpB) and variants of P(rbcL), and a version of the promoter P(trc) with two operators for enhanced repression, were developed and characterized in Synechocystis sp. strain PCC6803. (iv) It was shown that a system for targeted protein degradation, which is needed to enable dynamic expression studies, is working in Synechocystis sp. strain PCC6803. The pPMQAK1 shuttle vector allows the use of the growing numbers of BioBrick parts in many prokaryotes, and the other tools herein implemented facilitate the development of new parts and systems in cyanobacteria.
Figures





Similar articles
-
A chimeric vector for dual use in cyanobacteria and Escherichia coli, tested with cystatin, a nonfluorescent reporter protein.PeerJ. 2021 Nov 3;9:e12199. doi: 10.7717/peerj.12199. eCollection 2021. PeerJ. 2021. PMID: 34760347 Free PMC article.
-
A promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803.Gene. 1989 Dec 14;84(2):257-66. doi: 10.1016/0378-1119(89)90499-x. Gene. 1989. PMID: 2515116
-
Design and characterization of a synthetic minimal promoter for heterocyst-specific expression in filamentous cyanobacteria.PLoS One. 2018 Sep 11;13(9):e0203898. doi: 10.1371/journal.pone.0203898. eCollection 2018. PLoS One. 2018. PMID: 30204806 Free PMC article.
-
Synthetic Gene Regulation in Cyanobacteria.Adv Exp Med Biol. 2018;1080:317-355. doi: 10.1007/978-981-13-0854-3_13. Adv Exp Med Biol. 2018. PMID: 30091101 Review.
-
The current situations and limitations of genetic engineering in cyanobacteria: a mini review.Mol Biol Rep. 2023 Jun;50(6):5481-5487. doi: 10.1007/s11033-023-08456-8. Epub 2023 Apr 29. Mol Biol Rep. 2023. PMID: 37119415 Review.
Cited by
-
Biosafety of biotechnologically important microalgae: intrinsic suicide switch implementation in cyanobacterium Synechocystis sp. PCC 6803.Biol Open. 2016 Apr 15;5(4):519-28. doi: 10.1242/bio.017129. Biol Open. 2016. PMID: 27029902 Free PMC article.
-
Regulatory Tools for Controlling Gene Expression in Cyanobacteria.Adv Exp Med Biol. 2018;1080:281-315. doi: 10.1007/978-981-13-0854-3_12. Adv Exp Med Biol. 2018. PMID: 30091100 Free PMC article. Review.
-
CRISPR-Cas9/Cas12a biotechnology and application in bacteria.Synth Syst Biotechnol. 2018 Oct 3;3(3):135-149. doi: 10.1016/j.synbio.2018.09.004. eCollection 2018 Sep. Synth Syst Biotechnol. 2018. PMID: 30345399 Free PMC article. Review.
-
The effect of modulating the quantity of enzymes in a model ethanol pathway on metabolic flux in Synechocystis sp. PCC 6803.PeerJ. 2019 Aug 28;7:e7529. doi: 10.7717/peerj.7529. eCollection 2019. PeerJ. 2019. PMID: 31523505 Free PMC article.
-
New Applications of Synthetic Biology Tools for Cyanobacterial Metabolic Engineering.Front Bioeng Biotechnol. 2019 Feb 27;7:33. doi: 10.3389/fbioe.2019.00033. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 30873404 Free PMC article. Review.
References
-
- Chisti Y. Microalgae as sustainable cell factories. Environ. Engineer. Management J. 2006;5:261–274.
-
- Tamagnini P, Leitao E, Oliveira P, Ferreira D, Pinto F, Harris DJ, Heidorn T, Lindblad P. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol. Rev. 2007;31:692–720. - PubMed
-
- Koksharova O, Wolk C. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 2002;58:123–137. - PubMed
-
- Ow SY, Wright PC. Current trends in high throughput proteomics in cyanobacteria. FEBS Lett. 2009;583:1744–1752. - PubMed
-
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

