Development and evaluation of oligonucleotide chip based on the 16S-23S rRNA gene spacer region for detection of pathogenic microorganisms associated with sepsis
- PMID: 20237100
- PMCID: PMC2863927
- DOI: 10.1128/JCM.01130-09
Development and evaluation of oligonucleotide chip based on the 16S-23S rRNA gene spacer region for detection of pathogenic microorganisms associated with sepsis
Abstract
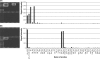
Oligonucleotide chips targeting the bacterial internal transcribed spacer region (ITS) of the 16S-23S rRNA gene, which contains genus- and species-specific regions, were developed and evaluated. Forty-three sequences were designed consisting of 1 universal, 3 Gram stain-specific, 9 genus-specific, and 30 species-specific probes. The specificity of the probes was confirmed using bacterial type strains including 54 of 52 species belonging to 18 genera. The performance of the probes was evaluated using 825 consecutive samples that were positive by blood culture in broth medium. Among the 825 clinical specimens, 708 (85.8%) were identified correctly by the oligonucleotide chip. Most (536 isolates, or 75.7%) were identified as staphylococci, Escherichia coli, or Klebsiella pneumoniae. Thirty-seven isolates (4.5%) did not bind to the corresponding specific probes. Most of these also were staphylococci, E. coli, or K. pneumoniae and accounted for 6.3% of total number of the species. Sixty-two specimens (7.5%) did not bind the genus- or species-specific probes because of lack of corresponding specific probes. Among them, Acinetobacter baumannii was the single most frequent isolate (26/62). The oligonucleotide chip was highly specific and sensitive in detecting the causative agents of bacteremia directly from positive blood cultures.
Figures
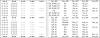
References
-
- Abelson, J. A., T. Moore, D. Bruckner, J. Deville, and K. Nielsen. 2005. Frequency of fungemia in hospitalized pediatric inpatients over 11 years at a tertiary care institution. Pediatrics 116:61-67. - PubMed
-
- Chung, J. W., H. S. Jeon, H. Sung, and M. N. Kim. 2009. Evaluation of MicroScan and Phoenix system for rapid identification and susceptibility testing using direct inoculation from positive BACTEC blood culture bottles. Korean J. Lab. Med. 29:25-34. - PubMed
-
- Fluit, A. C., F. J. Schmitz, and J. Verhoef. 2001. Frequency of isolation of pathogens from bloodstream, nosocomial pneumonia, skin and soft tissue, and urinary tract infections occurring in European patients. Eur. J. Clin. Microbiol. Infect. Dis. 20:188-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

