Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis
- PMID: 20237156
- PMCID: PMC2861616
- DOI: 10.1091/mbc.e09-07-0561
Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis
Abstract
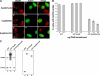
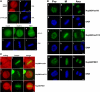
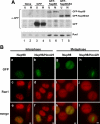
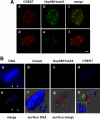
Chromosomal translocations involving the Nup98 gene are implicated in leukemias, especially acute myelogenous leukemia. These translocations generate chimeric fusion proteins, all of which have in common the N-terminal half of Nup98, which contains the nucleoporin FG/GLFG repeat motifs. The homeodomain group of Nup98 fusion proteins retain the C-terminus of a homeodomain transcription factor, including the homeobox responsible for DNA binding. Current models for Nup98 leukemogenesis invoke aberrant transcription resulting from recruitment of coregulators by the Nup98 repeat domain. Here we have investigated the behavior of Nup98-homeodomain fusion proteins throughout the cell cycle. At all stages, the fusion proteins exhibit a novel localization distinct from the component proteins or fragments. During interphase, there are dynamic interactions between the Nup98 fusions and endogenous Nup98 that lead to mislocalization of the intranuclear fraction of Nup98, but do not alter the level of Nup98 at the nuclear pore complex. During mitosis, no interaction between the fusion proteins and endogenous Nup98 is observed. However, the fusions are entirely concentrated at kinetochores and on chromosome arms, sites where the APC/C, a target of Nup98 regulation, is also found. Our observations suggest new possibilities for misregulation by which Nup98 translocations may contribute to cellular transformation and leukemogenesis.
Figures



Similar articles
-
Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype.PLoS One. 2016 Mar 31;11(3):e0152321. doi: 10.1371/journal.pone.0152321. eCollection 2016. PLoS One. 2016. PMID: 27031510 Free PMC article.
-
RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis.Cell Cycle. 2011 May 1;10(9):1456-67. doi: 10.4161/cc.10.9.15494. Epub 2011 May 1. Cell Cycle. 2011. PMID: 21467841
-
NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia.Cancer Res. 1998 Oct 1;58(19):4269-73. Cancer Res. 1998. PMID: 9766650
-
NUP98 gene fusions in hematologic malignancies.Leukemia. 2001 Nov;15(11):1689-95. doi: 10.1038/sj.leu.2402269. Leukemia. 2001. PMID: 11681408 Review.
-
NUP98 fusion in human leukemia: dysregulation of the nuclear pore and homeodomain proteins.Int J Hematol. 2005 Jul;82(1):21-7. doi: 10.1532/IJH97.04160. Int J Hematol. 2005. PMID: 16105755 Review.
Cited by
-
Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach.Cells. 2020 Jul 10;9(7):1666. doi: 10.3390/cells9071666. Cells. 2020. PMID: 32664447 Free PMC article.
-
Nuclear pore complexes: guardians of the nuclear genome.Cold Spring Harb Symp Quant Biol. 2010;75:585-97. doi: 10.1101/sqb.2010.75.059. Epub 2011 Apr 18. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21502404 Free PMC article. Review.
-
Ribonucleic Acid Export 1 Is a Kinetochore-Associated Protein That Participates in Chromosome Alignment in Mouse Oocytes.Int J Mol Sci. 2021 May 3;22(9):4841. doi: 10.3390/ijms22094841. Int J Mol Sci. 2021. PMID: 34063622 Free PMC article.
-
Loss of NOL10 leads to impaired disease progression of NUP98::DDX10 leukemia.Leukemia. 2025 Jun;39(6):1368-1379. doi: 10.1038/s41375-025-02607-5. Epub 2025 Apr 22. Leukemia. 2025. PMID: 40263434
-
Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic Transformation.Cancer Discov. 2022 Apr 1;12(4):1152-1169. doi: 10.1158/2159-8290.CD-21-0674. Cancer Discov. 2022. PMID: 34903620 Free PMC article.
References
-
- Argiropoulos B., Humphries R. K. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. - PubMed
-
- Bai X. T., Gu B. W., Yin T., Niu C., Xi X. D., Zhang J., Chen Z., Chen S. J. Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 2006;66:4584–4590. - PubMed
-
- Blevins M. B., Smith A. M., Phillips E. M., Powers M. A. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J. Biol. Chem. 2003;278:20979–20988. - PubMed
-
- Brown J. A., Bharathi A., Ghosh A., Whalen W., Fitzgerald E., Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in Poly(A+) RNA export and in the cytoskeleton. J. Biol. Chem. 1995;270:7411–7419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

