The 74-kilodalton immunodominant antigen of the pathogenic oomycete Pythium insidiosum is a putative exo-1,3-beta-glucanase
- PMID: 20237199
- PMCID: PMC2916248
- DOI: 10.1128/CVI.00515-09
The 74-kilodalton immunodominant antigen of the pathogenic oomycete Pythium insidiosum is a putative exo-1,3-beta-glucanase
Abstract
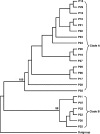
The oomycetous, fungus-like, aquatic organism Pythium insidiosum is the causative agent of pythiosis, a life-threatening infectious disease of humans and animals living in tropical and subtropical areas of the world. Common sites of infection are the arteries, eyes, cutaneous/subcutaneous tissues, and gastrointestinal tract. Diagnosis of pythiosis is time-consuming and difficult. Radical excision of the infected organs is the main treatment for pythiosis because conventional antifungal drugs are ineffective. An immunotherapeutic vaccine prepared from P. insidiosum crude extract showed limited efficacy in the treatment of pythiosis patients. Many pythiosis patients suffer lifelong disabilities or die from an advanced infection. Recently, we identified a 74-kDa major immunodominant antigen of P. insidiosum which could be a target for development of a more effective serodiagnostic test and vaccines. Mass spectrometric analysis identified two peptides of the 74-kDa antigen (s74-1 and s74-2) which perfectly matched a putative exo-1,3-ss-glucanase (EXO1) of Phytophthora infestans. Using degenerate primers derived from these peptides, a 1.1-kb product was produced by PCR, and its sequence was found to be homologous to that of the P. infestans exo-1,3-ss-glucanase gene, EXO1. Enzyme-linked immunosorbent assays targeting the s74-1 and s74-2 synthetic peptides demonstrated that the 74-kDa antigen was highly immunoreactive with pythiosis sera but not with control sera. Phylogenetic analysis using part of the 74-kDa protein-coding sequence divided 22 Thai isolates of P. insidiosum into two clades. Further characterization of the putative P. insidiosum glucanase could lead to new diagnostic tests and to antimicrobial agents and vaccines for the prevention and management of the serious and life-threatening disease of pythiosis.
Figures
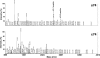

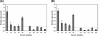
Similar articles
-
The Immunoreactive Exo-1,3-β-Glucanase from the Pathogenic Oomycete Pythium insidiosum Is Temperature Regulated and Exhibits Glycoside Hydrolase Activity.PLoS One. 2015 Aug 11;10(8):e0135239. doi: 10.1371/journal.pone.0135239. eCollection 2015. PLoS One. 2015. PMID: 26263509 Free PMC article.
-
Expression, purification, and characterization of the recombinant exo-1,3-β-glucanase (Exo1) of the pathogenic oomycete Pythium insidiosum.Heliyon. 2020 Jun 19;6(6):e04237. doi: 10.1016/j.heliyon.2020.e04237. eCollection 2020 Jun. Heliyon. 2020. PMID: 32596527 Free PMC article.
-
Identification of a novel 74-kiloDalton immunodominant antigen of Pythium insidiosum recognized by sera from human patients with pythiosis.J Clin Microbiol. 2006 May;44(5):1674-80. doi: 10.1128/JCM.44.5.1674-1680.2006. J Clin Microbiol. 2006. PMID: 16672392 Free PMC article.
-
Review of methods and antimicrobial agents for susceptibility testing against Pythium insidiosum.Heliyon. 2020 Apr 12;6(4):e03737. doi: 10.1016/j.heliyon.2020.e03737. eCollection 2020 Apr. Heliyon. 2020. PMID: 32322727 Free PMC article. Review.
-
History and Perspective of Immunotherapy for Pythiosis.Vaccines (Basel). 2021 Sep 26;9(10):1080. doi: 10.3390/vaccines9101080. Vaccines (Basel). 2021. PMID: 34696188 Free PMC article. Review.
Cited by
-
Automated Cell-Free Multiprotein Synthesis Facilitates the Identification of a Secretory, Oligopeptide Elicitor-Like, Immunoreactive Protein of the Oomycete Pythium insidiosum.mSystems. 2020 May 12;5(3):e00196-20. doi: 10.1128/mSystems.00196-20. mSystems. 2020. PMID: 32398276 Free PMC article.
-
The Immunoreactive Exo-1,3-β-Glucanase from the Pathogenic Oomycete Pythium insidiosum Is Temperature Regulated and Exhibits Glycoside Hydrolase Activity.PLoS One. 2015 Aug 11;10(8):e0135239. doi: 10.1371/journal.pone.0135239. eCollection 2015. PLoS One. 2015. PMID: 26263509 Free PMC article.
-
Expression, purification, and characterization of the recombinant exo-1,3-β-glucanase (Exo1) of the pathogenic oomycete Pythium insidiosum.Heliyon. 2020 Jun 19;6(6):e04237. doi: 10.1016/j.heliyon.2020.e04237. eCollection 2020 Jun. Heliyon. 2020. PMID: 32596527 Free PMC article.
-
Prospecting Biomarkers for Diagnostic and Therapeutic Approaches in Pythiosis.J Fungi (Basel). 2021 May 28;7(6):423. doi: 10.3390/jof7060423. J Fungi (Basel). 2021. PMID: 34071174 Free PMC article.
References
-
- Abuhammour, W., and E. Habte-Gaber. 2004. Newer antifungal agents. Indian J. Pediatr. 71:253-259. - PubMed
-
- Adams, D. J. 2004. Fungal cell wall chitinases and glucanases. Microbiology 150:2029-2035. - PubMed
-
- Brown, T. A., A. M. Grooters, and G. L. Hosgood. 2008. In vitro susceptibility of Pythium insidiosum and a Lagenidium sp to itraconazole, posaconazole, voriconazole, terbinafine, caspofungin, and mefenoxam. Am. J. Vet. Res. 69:1463-1468. - PubMed
-
- Brown, C. C., J. J. McClure, P. Triche, and C. Crowder. 1988. Use of immunohistochemical methods for diagnosis of equine pythiosis. Am. J. Vet. Res. 49:1866-1868. - PubMed
-
- Chaiprasert, A., T. Krajaejun, S. Pannanusorn, C. Prariyachatigul, W. Wanachiwanawin, B. Sathapatayavongs, T. Juthayothin, N. Smittipat, N. Vanittanakom, and A. Chindamporn. 2009. Pythium insidiosum Thai isolates: molecular phylogenetic analysis. Asian Biomed. 3:623-633.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

