Accelerated evolution of PAK3- and PIM1-like kinase gene families in the zebra finch, Taeniopygia guttata
- PMID: 20237222
- PMCID: PMC3889628
- DOI: 10.1093/molbev/msq080
Accelerated evolution of PAK3- and PIM1-like kinase gene families in the zebra finch, Taeniopygia guttata
Abstract
Genes encoding protein kinases tend to evolve slowly over evolutionary time, and only rarely do they appear as recent duplications in sequenced vertebrate genomes. Consequently, it was a surprise to find two families of kinase genes that have greatly and recently expanded in the zebra finch (Taeniopygia guttata) lineage. In contrast to other amniotic genomes (including chicken) that harbor only single copies of p21-activated serine/threonine kinase 3 (PAK3) and proviral integration site 1 (PIM1) genes, the zebra finch genome appeared at first to additionally contain 67 PAK3-like (PAK3L) and 51 PIM1-like (PIM1L) protein kinase genes. An exhaustive analysis of these gene models, however, revealed most to be incomplete, owing to the absence of terminal exons. After reprediction, 31 PAK3L genes and 10 PIM1L genes remain, and all but three are predicted, from the retention of functional sites and open reading frames, to be enzymatically active. PAK3L, but not PIM1L, gene sequences show evidence of recurrent episodes of positive selection, concentrated within structures spatially adjacent to N- and C-terminal protein regions that have been discarded from zebra finch PAK3L genes. At least seven zebra finch PAK3L genes were observed to be expressed in testis, whereas two sequences were found transcribed in the brain, one broadly including the song nuclei and the other in the ventricular zone and in cells resembling Bergmann's glia in the cerebellar Purkinje cell layer. Two PIM1L sequences were also observed to be expressed with broad distributions in the zebra finch brain, one in both the ventricular zone and the cerebellum and apparently associated with glial cells and the other showing neuronal cell expression and marked enrichment in midbrain/thalamic nuclei. These expression patterns do not correlate with zebra finch-specific features such as vocal learning. Nevertheless, our results show how ancient and conserved intracellular signaling molecules can be co-opted, following duplication, thereby resulting in lineage-specific functions, presumably affecting the zebra finch testis and brain.
Figures

 (nonsynonymous/synonymous substitution rates ratio) and divergence times (in Mya) for the clade including all PAK3L genes, Clades 1 and 2 are shown.
(nonsynonymous/synonymous substitution rates ratio) and divergence times (in Mya) for the clade including all PAK3L genes, Clades 1 and 2 are shown.

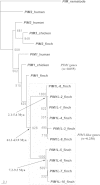
 for PIM1L genes and other PIM1 genes are provided. The estimated timescales for the duplications of PIM1L genes are shown (in Mya).
for PIM1L genes and other PIM1 genes are provided. The estimated timescales for the duplications of PIM1L genes are shown (in Mya).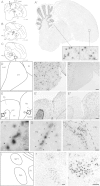
Similar articles
-
Karyotypic polymorphism of the zebra finch Z chromosome.Chromosoma. 2011 Jun;120(3):255-64. doi: 10.1007/s00412-010-0308-3. Epub 2011 Jan 11. Chromosoma. 2011. PMID: 21369954 Free PMC article.
-
The zebra finch neuropeptidome: prediction, detection and expression.BMC Biol. 2010 Apr 1;8:28. doi: 10.1186/1741-7007-8-28. BMC Biol. 2010. PMID: 20359331 Free PMC article.
-
Genomic and neural analysis of the estradiol-synthetic pathway in the zebra finch.BMC Neurosci. 2010 Apr 1;11:46. doi: 10.1186/1471-2202-11-46. BMC Neurosci. 2010. PMID: 20359328 Free PMC article.
-
Identification, localisation and functional implication of 26RFa orthologue peptide in the brain of zebra finch (Taeniopygia guttata).J Neuroendocrinol. 2011 Sep;23(9):791-803. doi: 10.1111/j.1365-2826.2011.02179.x. J Neuroendocrinol. 2011. PMID: 21696471
-
The zebra finch, Taeniopygia guttata: an avian model for investigating the neurobiological basis of vocal learning.Cold Spring Harb Protoc. 2014 Oct 23;2014(12):1237-42. doi: 10.1101/pdb.emo084574. Cold Spring Harb Protoc. 2014. PMID: 25342070 Free PMC article. Review.
Cited by
-
Micro Germline-Restricted Chromosome in Blue Tits: Evidence for Meiotic Functions.Mol Biol Evol. 2023 May 2;40(5):msad096. doi: 10.1093/molbev/msad096. Mol Biol Evol. 2023. PMID: 37116210 Free PMC article.
-
Avian genomics lends insights into endocrine function in birds.Gen Comp Endocrinol. 2018 Jan 15;256:123-129. doi: 10.1016/j.ygcen.2017.05.023. Epub 2017 Jun 17. Gen Comp Endocrinol. 2018. PMID: 28596079 Free PMC article. Review.
-
Karyotypic polymorphism of the zebra finch Z chromosome.Chromosoma. 2011 Jun;120(3):255-64. doi: 10.1007/s00412-010-0308-3. Epub 2011 Jan 11. Chromosoma. 2011. PMID: 21369954 Free PMC article.
-
The PAKs come of age: Celebrating 18 years of discovery.Cell Logist. 2012 Apr 1;2(2):54-58. doi: 10.4161/cl.22084. Cell Logist. 2012. PMID: 23125949 Free PMC article.
-
Using the canary genome to decipher the evolution of hormone-sensitive gene regulation in seasonal singing birds.Genome Biol. 2015 Jan 29;16(1):19. doi: 10.1186/s13059-014-0578-9. Genome Biol. 2015. PMID: 25631560 Free PMC article.
References
-
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

