Genetic analysis of variation in transcription factor binding in yeast
- PMID: 20237471
- PMCID: PMC2941147
- DOI: 10.1038/nature08934
Genetic analysis of variation in transcription factor binding in yeast
Abstract
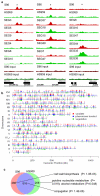
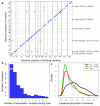
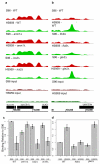
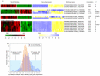
Variation in transcriptional regulation is thought to be a major cause of phenotypic diversity. Although widespread differences in gene expression among individuals of a species have been observed, studies to examine the variability of transcription factor binding on a global scale have not been performed, and thus the extent and underlying genetic basis of transcription factor binding diversity is unknown. By mapping differences in transcription factor binding among individuals, here we present the genetic basis of such variation on a genome-wide scale. Whole-genome Ste12-binding profiles were determined using chromatin immunoprecipitation coupled with DNA sequencing in pheromone-treated cells of 43 segregants of a cross between two highly diverged yeast strains and their parental lines. We identified extensive Ste12-binding variation among individuals, and mapped underlying cis- and trans-acting loci responsible for such variation. We showed that most transcription factor binding variation is cis-linked, and that many variations are associated with polymorphisms residing in the binding motifs of Ste12 as well as those of several proposed Ste12 cofactors. We also identified two trans-factors, AMN1 and FLO8, that modulate Ste12 binding to promoters of more than ten genes under alpha-factor treatment. Neither of these two genes was previously known to regulate Ste12, and we suggest that they may be mediators of gene activity and phenotypic diversity. Ste12 binding strongly correlates with gene expression for more than 200 genes, indicating that binding variation is functional. Many of the variable-bound genes are involved in cell wall organization and biogenesis. Overall, these studies identified genetic regulators of molecular diversity among individuals and provide new insights into mechanisms of gene regulation.
Figures

References
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188(4184):107. - PubMed
-
- Pennisi E. Searching for the genome's second code. Science. 2004;306(5696):632. - PubMed
-
- Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422(6929):297. - PubMed
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296(5568):752. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

