Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium
- PMID: 20237561
- PMCID: PMC3048781
- DOI: 10.1038/nature08850
Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium
Abstract
Fusarium species are among the most important phytopathogenic and toxigenic fungi. To understand the molecular underpinnings of pathogenicity in the genus Fusarium, we compared the genomes of three phenotypically diverse species: Fusarium graminearum, Fusarium verticillioides and Fusarium oxysporum f. sp. lycopersici. Our analysis revealed lineage-specific (LS) genomic regions in F. oxysporum that include four entire chromosomes and account for more than one-quarter of the genome. LS regions are rich in transposons and genes with distinct evolutionary profiles but related to pathogenicity, indicative of horizontal acquisition. Experimentally, we demonstrate the transfer of two LS chromosomes between strains of F. oxysporum, converting a non-pathogenic strain into a pathogen. Transfer of LS chromosomes between otherwise genetically isolated strains explains the polyphyletic origin of host specificity and the emergence of new pathogenic lineages in F. oxysporum. These findings put the evolution of fungal pathogenicity into a new perspective.
Figures


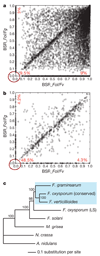
Comment in
-
Expanding horizons through chromosome exchange.Nat Rev Microbiol. 2010 May;8(5):311. doi: 10.1038/nrmicro2357. Nat Rev Microbiol. 2010. PMID: 21080586 No abstract available.
References
-
- Agrios GN. Plant Pathology. 5th edn. Academic Press; 2005.
-
- Armstrong GM, Armstrong JK. In: Fusarium: Diseases, Biology and Taxonomy. Nelson PE, Toussoun TA, Cook R, editors. Penn State University Press; 1981. pp. 391–399.
-
- O’Donnell K, et al. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 2004;42:5109–5120. - PMC - PubMed
-
- Cuomo CA, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

