Codominant scoring of AFLP in association panels
- PMID: 20237752
- PMCID: PMC2886132
- DOI: 10.1007/s00122-010-1313-x
Codominant scoring of AFLP in association panels
Abstract
A study on the codominant scoring of AFLP markers in association panels without prior knowledge on genotype probabilities is described. Bands are scored codominantly by fitting normal mixture models to band intensities, illustrating and optimizing existing methodology, which employs the EM-algorithm. We study features that improve the performance of the algorithm, and the unmixing in general, like parameter initialization, restrictions on parameters, data transformation, and outlier removal. Parameter restrictions include equal component variances, equal or nearly equal distances between component means, and mixing probabilities according to Hardy-Weinberg Equilibrium. Histogram visualization of band intensities with superimposed normal densities, and optional classification scores and other grouping information, assists further in the codominant scoring. We find empirical evidence favoring the square root transformation of the band intensity, as was found in segregating populations. Our approach provides posterior genotype probabilities for marker loci. These probabilities can form the basis for association mapping and are more useful than the standard scoring categories A, H, B, C, D. They can also be used to calculate predictors for additive and dominance effects. Diagnostics for data quality of AFLP markers are described: preference for three-component mixture model, good separation between component means, and lack of singletons for the component with highest mean. Software has been developed in R, containing the models for normal mixtures with facilitating features, and visualizations. The methods are applied to an association panel in tomato, comprising 1,175 polymorphic markers on 94 tomato hybrids, as part of a larger study within the Dutch Centre for BioSystems Genomics.
Figures
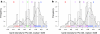


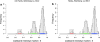
References
-
- van Berloo R, van Heusden S, Bovy A, Meijer-Dekens F, Lindhout P, van Eeuwijk F. Genetic research in a public–private research consortium: prospects for indirect use of Elite breeding germplasm in academic research. Euphytica. 2008;161:293–300. doi: 10.1007/s10681-007-9519-y. - DOI
-
- Bezdek J. Pattern recognition with fuzzy objective function algorithms. New York: Plenum Press; 1981.
-
- Böhning D, Seidel W, Alf M, Garel B, Patilea V, Walther G. Advances in mixture models. Comput Stat Data Anal. 2007;51:5205–5210. doi: 10.1016/j.csda.2006.10.025. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

