Human LPLUNC1 is a secreted product of goblet cells and minor glands of the respiratory and upper aerodigestive tracts
- PMID: 20237794
- PMCID: PMC2852594
- DOI: 10.1007/s00418-010-0683-0
Human LPLUNC1 is a secreted product of goblet cells and minor glands of the respiratory and upper aerodigestive tracts
Abstract
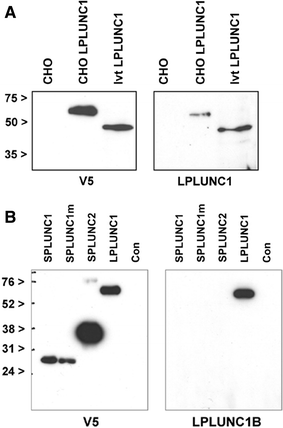
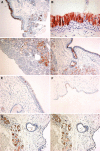
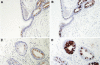
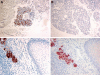
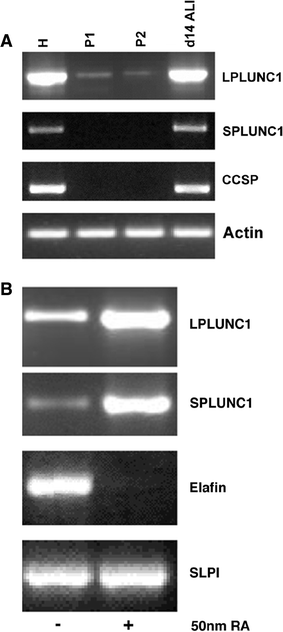
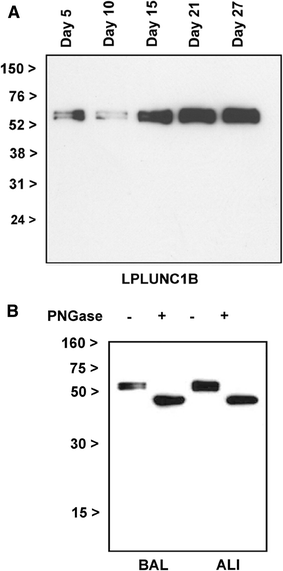
Long PLUNC1 (LPLUNC1, C20orf114) is a member of a family of poorly described proteins (PLUNCS) expressed in the upper respiratory tract and oral cavity, which may function in host defence. Although it is one of the most highly expressed genes in the upper airways and has been identified in sputum and nasal secretions by proteomic studies, localisation of LPLUNC1 protein has not yet been described. We developed affinity purified antibodies and localised the protein in tissues of the human respiratory tract, oro- and nasopharynx. We have complemented these studies with analysis of LPLUNC1 expression in primary human lung cell cultures and used Western blotting to study the protein in cell culture secretions and in BAL. LPLUNC1 is a product of a population of goblet cells in the airway epithelium and nasal passages and is also present in airway submucosal glands and minor glands of the oral and nasal cavities. The protein is not expressed in peripheral lung epithelial cells. LPLUNC1 is present in bronchoalveolar lavage fluid as two glycosylated isoforms and primary airway epithelial cells produce identical proteins as they undergo mucociliary differentiation. Our results suggest that LPLUNC1 is an abundant, secreted product of goblet cells and minor mucosal glands of the respiratory tract and oral cavity and suggest that the protein functions in the complex milieu that protects the mucosal surfaces in these locations.
Figures

References
-
- Bingle CD, Bingle L. Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta. 2000;1493:363–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

