Annexin A4 interacts with the NF-kappaB p50 subunit and modulates NF-kappaB transcriptional activity in a Ca2+-dependent manner
- PMID: 20237821
- PMCID: PMC11115496
- DOI: 10.1007/s00018-010-0331-9
Annexin A4 interacts with the NF-kappaB p50 subunit and modulates NF-kappaB transcriptional activity in a Ca2+-dependent manner
Abstract
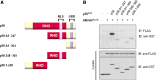
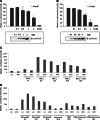
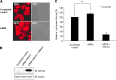
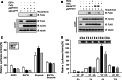
Previously, we identified annexin A4 (ANXA4) as a candidate substrate of caspase-3. Proteomic studies were performed to identify interacting proteins with a view to determining the roles of ANXA4. ANXA4 was found to interact with the p105. Subsequent studies revealed that ANXA4 interacts with NF-kappaB through the Rel homology domain of p50. Furthermore, the interaction is markedly increased by elevated Ca(2+) levels. NF-kappaB transcriptional activity assays demonstrated that ANXA4 suppresses NF-kappaB transcriptional activity in the resting state. Following treatment with TNF-alpha or PMA, ANXA4 also suppressed NF-kappaB transcriptional activity, which was upregulated significantly early after etoposide treatment. This difference may be due to the intracellular Ca(2+) level. Additionally, ANXA4 translocates to the nucleus together with p50, and imparts greater resistance to apoptotic stimulation by etoposide. Our results collectively indicate that ANXA4 differentially modulates the NF-kappaB signaling pathway, depending on its interactions with p50 and the intracellular Ca(2+) ion level.
Figures


Similar articles
-
p53 and ANXA4/NF‑κB p50 complexes regulate cell proliferation, apoptosis and tumor progression in ovarian clear cell carcinoma.Int J Mol Med. 2020 Dec;46(6):2102-2114. doi: 10.3892/ijmm.2020.4757. Epub 2020 Oct 14. Int J Mol Med. 2020. PMID: 33125094 Free PMC article.
-
Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-κB activation by interacting with p65/RelA and p50/NF-κB1.J Virol. 2013 Dec;87(23):12935-48. doi: 10.1128/JVI.01952-13. Epub 2013 Sep 25. J Virol. 2013. PMID: 24067962 Free PMC article.
-
Membrane-cytoplasm translocation of annexin A4 is involved in the metastasis of colorectal carcinoma.Aging (Albany NY). 2021 Mar 24;13(7):10312-10325. doi: 10.18632/aging.202793. Epub 2021 Mar 24. Aging (Albany NY). 2021. PMID: 33761465 Free PMC article.
-
The role of annexin A4 in cancer.Front Biosci (Landmark Ed). 2016 Jun 1;21(5):949-57. doi: 10.2741/4432. Front Biosci (Landmark Ed). 2016. PMID: 27100483 Review.
-
Annexin A4 and cancer.Clin Chim Acta. 2015 Jul 20;447:72-8. doi: 10.1016/j.cca.2015.05.016. Epub 2015 Jun 3. Clin Chim Acta. 2015. PMID: 26048190 Review.
Cited by
-
Intracellular annexin A2 regulates NF-κB signaling by binding to the p50 subunit: implications for gemcitabine resistance in pancreatic cancer.Cell Death Dis. 2015 Jan 22;6(1):e1606. doi: 10.1038/cddis.2014.558. Cell Death Dis. 2015. PMID: 25611381 Free PMC article.
-
Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells.Int J Mol Sci. 2020 Dec 29;22(1):252. doi: 10.3390/ijms22010252. Int J Mol Sci. 2020. PMID: 33383772 Free PMC article.
-
The transcription factor Ikaros inhibits cell proliferation by downregulating ANXA4 expression in hepatocellular carcinoma.Am J Cancer Res. 2017 Jun 1;7(6):1285-1297. eCollection 2017. Am J Cancer Res. 2017. PMID: 28670491 Free PMC article.
-
Muscadine Grape Skin Extract Induces an Unfolded Protein Response-Mediated Autophagy in Prostate Cancer Cells: A TMT-Based Quantitative Proteomic Analysis.PLoS One. 2016 Oct 18;11(10):e0164115. doi: 10.1371/journal.pone.0164115. eCollection 2016. PLoS One. 2016. PMID: 27755556 Free PMC article.
-
Proteomics Profiling of Autologous Blood and Semen Exosomes from HIV-infected and Uninfected Individuals Reveals Compositional and Functional Variabilities.Mol Cell Proteomics. 2020 Jan;19(1):78-100. doi: 10.1074/mcp.RA119.001594. Epub 2019 Nov 1. Mol Cell Proteomics. 2020. PMID: 31676584 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

