Pro-apoptotic and antiproliferative activity of human KCNRG, a putative tumor suppressor in 13q14 region
- PMID: 20237900
- PMCID: PMC2803748
- DOI: 10.1007/s13277-009-0005-0
Pro-apoptotic and antiproliferative activity of human KCNRG, a putative tumor suppressor in 13q14 region
Abstract
Deletion of 13q14.3 and a candidate gene KCNRG (potassium channel regulating gene) is the most frequent chromosomal abnormality in B-cell chronic lymphocytic leukemia and is a common finding in multiple myeloma (MM). KCNRG protein may interfere with the normal assembly of the K+ channel proteins causing the suppression of Kv currents. We aimed to examine possible role of KCNRG haploinsufficiency in chronic lymphocytic leukemia (CLL) and MM cells. We performed detailed genomic analysis of the KCNRG locus; studied effects of the stable overexpression of KCNRG isoforms in RPMI-8226, HL-60, and LnCaP cells; and evaluated relative expression of its transcripts in various human lymphomas. Three MM cell lines and 35 CLL PBL samples were screened for KCNRG mutations. KCNRG exerts growth suppressive and pro-apoptotic effects in HL-60, LnCaP, and RPMI-8226 cells. Direct sequencing of KCNRG exons revealed point mutation delT in RPMI-8226 cell line. Levels of major isoform of KCNRG mRNA are lower in DLBL lymphomas compared to normal PBL samples, while levels of its minor mRNA are decreased across the broad range of the lymphoma types. The haploinsufficiency of KCNRG might be relevant to the progression of CLL and MM at least in a subset of patients.
Figures
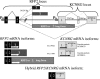



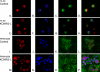


References
-
- Gale RP, Foon KA. Biology of chronic lymphocytic leukaemia. Semin Hematol. 1987;24:209–229. - PubMed
-
- Ougolkov AV, Bone ND, Fernandez-Zapico ME, Kay NE, Billadeau DD. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukaemia B cells. Blood. 2007;110:735–742. doi: 10.1182/blood-2006-12-060947. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

